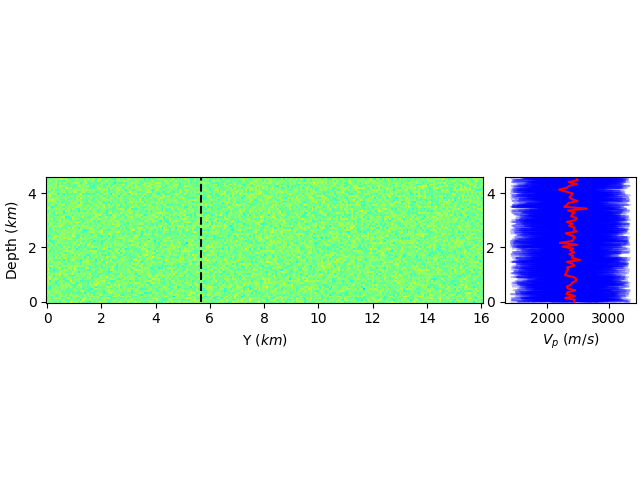
I would like to produce a figure where I display a 2d data on the left and a vertical slice of said data on the right.
I successfully did it with the following MWE:
import matplotlib.pyplot as plt
import numpy as np
nz = 66
nx = 130
ny = 230
h = 70
ensemble_size = 50
x = np.array([ix*h / 1000 for ix in range(nx)])
y = np.array([iy*h / 1000 for iy in range(ny)])
z = np.array([iz*h / 1000 for iz in range(nz)])
Y, Z = np.meshgrid(y, z, indexing='ij')
vp_min = 1400
vp_max = 3350
p_max = 1200
ix = 42
iy = 81
vp = (vp_max - vp_min)*np.random.rand(nz*nx*ny, ensemble_size) vp_min
vp_mean = np.mean(vp, axis=1).reshape(nz*nx*ny, 1)
########################################################################
fig, (ax1, ax2) = plt.subplots(ncols=2, sharey=True)
ax1.pcolor(Y, Z, vp_mean.reshape(nz, nx, ny, order='F')[:,ix,:].T, cmap="jet", vmin=vp_min, vmax=vp_max)
ax1.plot(iy*h*np.ones(nz) / 1000, z, "k--")
ax1.set_ylabel(r'Depth ($km$)')
ax1.set_xlabel(r'Y ($km$)')
ax1.set_aspect('equal')
lines = []
for e in range(ensemble_size):
lines.append( ax2.plot(vp[:,e].reshape(nz, nx, ny, order='F')[:,ix,iy], z, "b", alpha=0.25, label="models") )
lines.append( ax2.plot(vp_mean.reshape(nz, nx, ny, order='F')[:,ix,iy], z, "r", alpha=1.0, label="average model") )
plt.setp(lines[1:ensemble_size], label="_")
ax2.set_xlabel(r'$V_p$ ($m/s$)')
ax2.invert_yaxis()
ax2.legend()
########################################################################
plt.savefig("log_" str(ix) "_" str(iy) ".pdf", bbox_inches='tight')
plt.show(block=False)
plt.close("all")
However, I'm unsatisfied by the aspect ratio of my left sub-figure 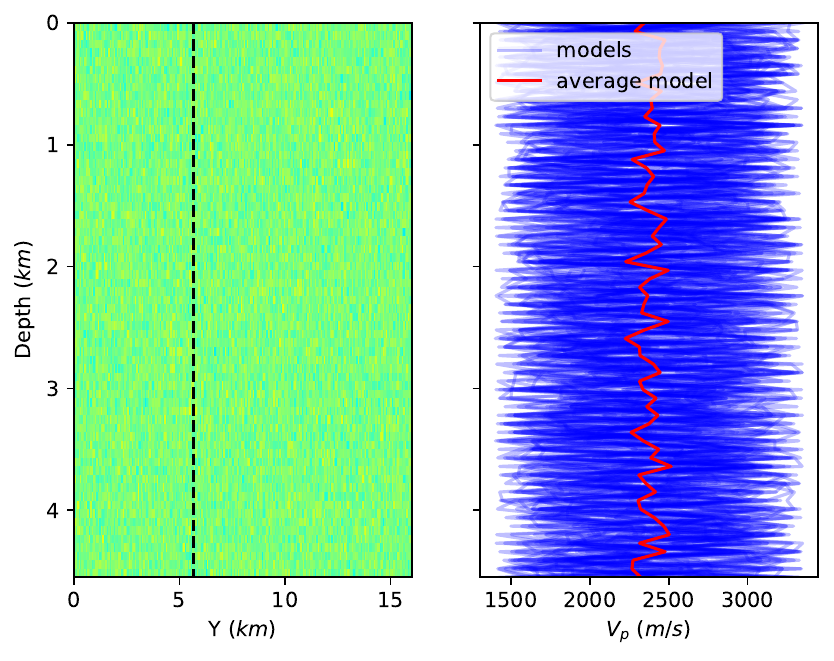 .
.
I would rather have the following layout 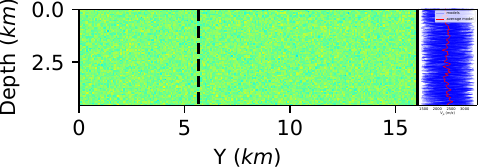 But I don't quite know how to do it.
But I don't quite know how to do it.
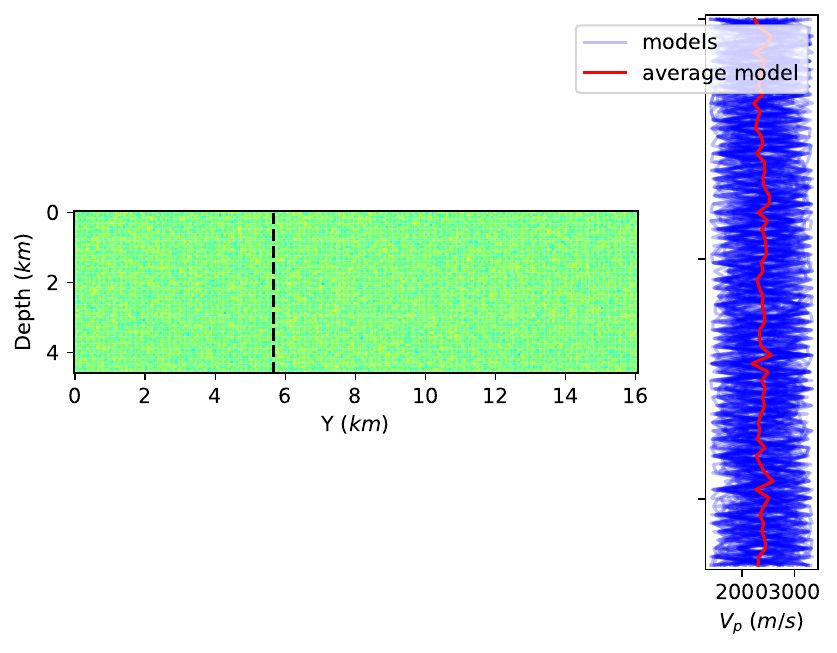
EDIT I also tried to use
fig, (ax1, ax2) = plt.subplots(ncols=2, sharey=True, gridspec_kw={'width_ratios':[5, 1]})
...
ax1.set_aspect(1)
However, the result isn't quite what I want as the two depth axes do not have the same size anymore:
CodePudding user response:
A second approach, aside from manually adjusting the figure size is to use an inset_axes that is a child of the parent. You can even set up sharing between the two, though you'll have to remove the tick labels if that is what you want:
fig, ax1 = plt.subplots(constrained_layout=True)
ax1.pcolor(Y, Z, vp_mean.reshape(nz, nx, ny, order='F')[:,ix,:].T, cmap="jet", vmin=vp_min, vmax=vp_max)
ax1.plot(iy*h*np.ones(nz) / 1000, z, "k--")
ax1.set_ylabel(r'Depth ($km$)')
ax1.set_xlabel(r'Y ($km$)')
ax1.set_aspect('equal')
ax2 = ax1.inset_axes([1.05, 0, 0.3, 1], transform=ax1.transAxes)
ax1.get_shared_y_axes().join(ax1, ax2)
lines = []
for e in range(ensemble_size):
lines.append( ax2.plot(vp[:,e].reshape(nz, nx, ny, order='F')[:,ix,iy], z, "b", alpha=0.25, label="models") )
lines.append( ax2.plot(vp_mean.reshape(nz, nx, ny, order='F')[:,ix,iy], z, "r", alpha=1.0, label="average model") )
plt.setp(lines[1:ensemble_size], label="_")
ax2.set_xlabel(r'$V_p$ ($m/s$)')
This has the advantage that the second axes will always be there, even if you zoom the first axes.