Possibly this question has been asked before but I don't manage to find a solution. I have a TSV (test.tsv) that looks like this:
TPM variants NOI_ID HLA NAL
324.28 several p1 one 2
169.21 NA p1 two 15
154.78 NA p1 three 0
143.31 NA p1 four 2
468.7 NA p2 five 0
322.76 several p2 six 2
620.98 NA p2 two 0
591.17 NA p2 seven 0
637.74 NA p3 eight 4
519.8 NA p3 nine 10
1439.58 NA p3 ten 23
122.05 NA p4 five 14
149.74 NA p4 eleven 77
213.7 NA p4 twelve 100
162.53 NA p4 one 15
Using this code:
fig13 <- read.table("test.tsv", sep="\t", header=T, check.names=FALSE)
fig13 = fig13[-which(grepl("_w8", fig13$NOI_ID)),]
fig13_melt <- melt(fig13[c("TPM", "NOI_ID", "HLA", "variants")], id=c("NOI_ID", "HLA", "variants"))
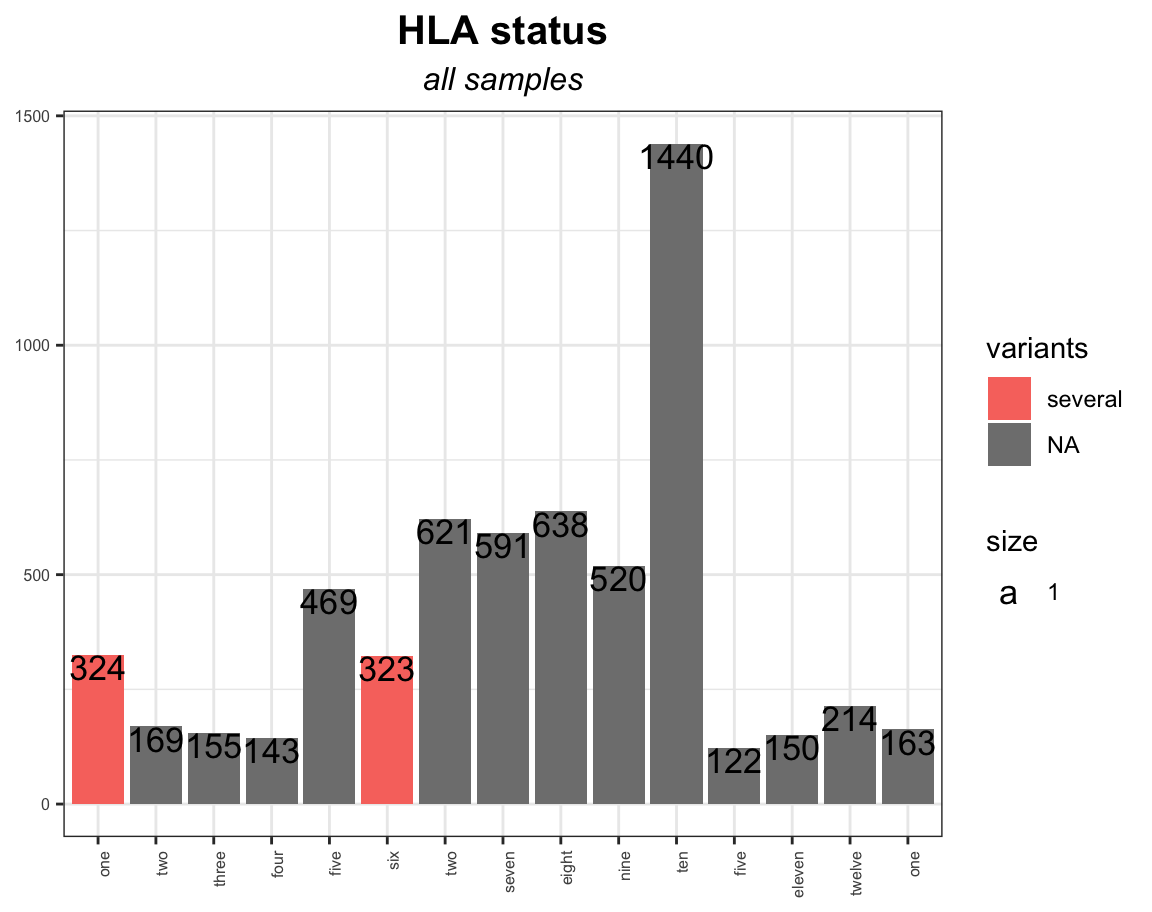
hla_barplot = ggplot(fig13_melt, aes(x=1:nrow(fig13_melt), y=value, fill=variants))
geom_bar(stat="identity", position="dodge") ggtitle("HLA status", subtitle = "all samples") theme_bw()
theme(axis.title.y=element_blank(), axis.title.x=element_blank(),
plot.title = element_text(face = "bold", size = 15, hjust = 0.5),
plot.subtitle=element_text(size=12, hjust=0.5, face="italic"),
axis.text.y = element_text(size=6),
axis.text.x = element_text(angle = 90, hjust=1, size=6))
geom_text(aes(label = round(value,0), vjust = 1, size = 1))
scale_x_discrete(labels=fig13_melt$HLA, breaks=1:nrow(fig13_melt), limits=factor(1:nrow(fig13_melt)), name='HLA')
hla_barplot
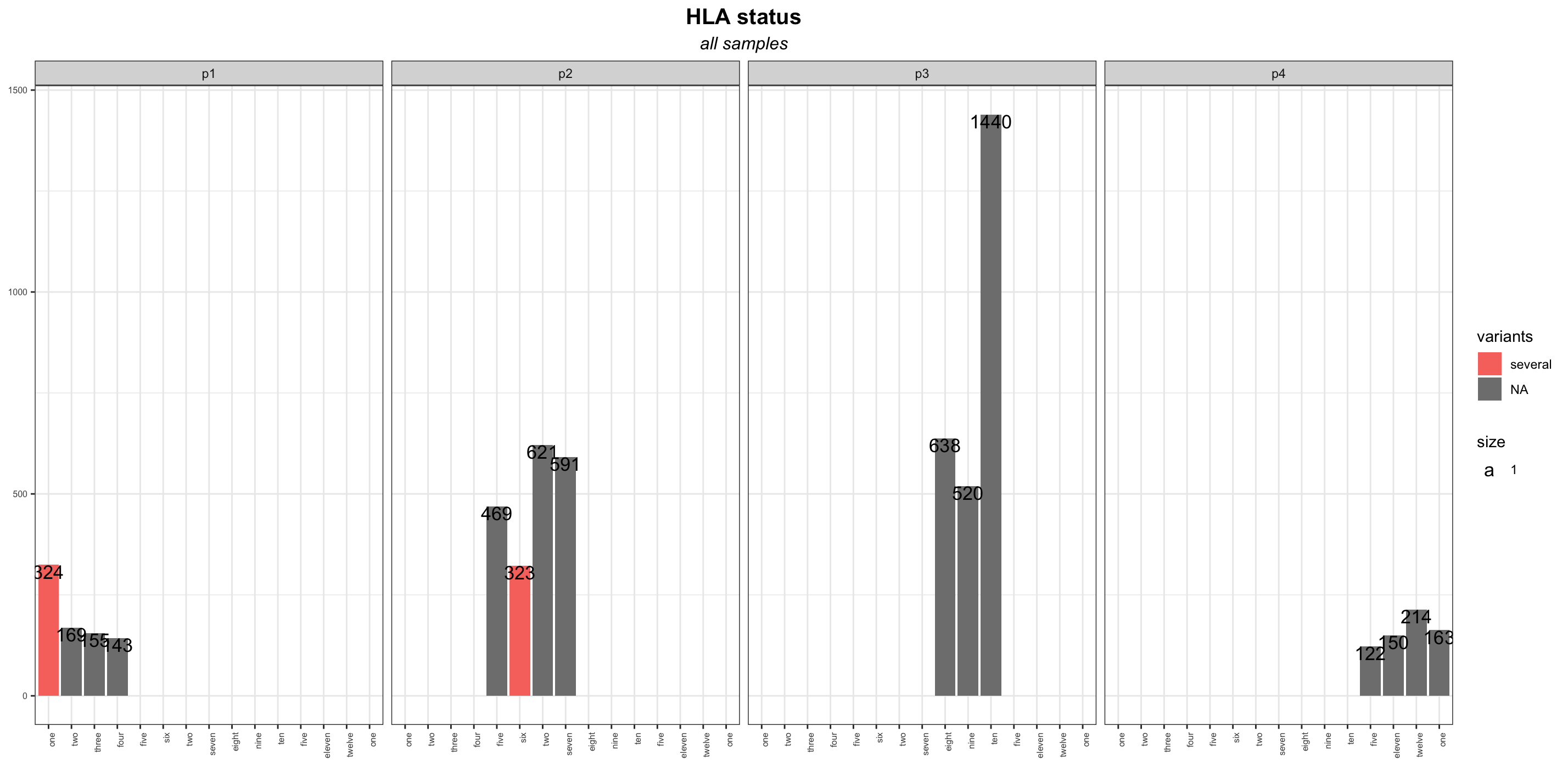
However when I try to use facet_wrap to split the plot:
hla_barplot facet_wrap(.~ NOI_ID, nrow = 1)
I get the following:
So, as far as I can see, each subplot has all the x labels of the whole dataset. Is there a way to just plot in the x-axis the labels of each subgroup?
CodePudding user response:
The issue is that you mapped 1:nrow(...) on x and fixed the limits of the x scale limits=factor(1:nrow(fig13_melt)). Doing so scales="free_x" as suggested in the comments will not work.
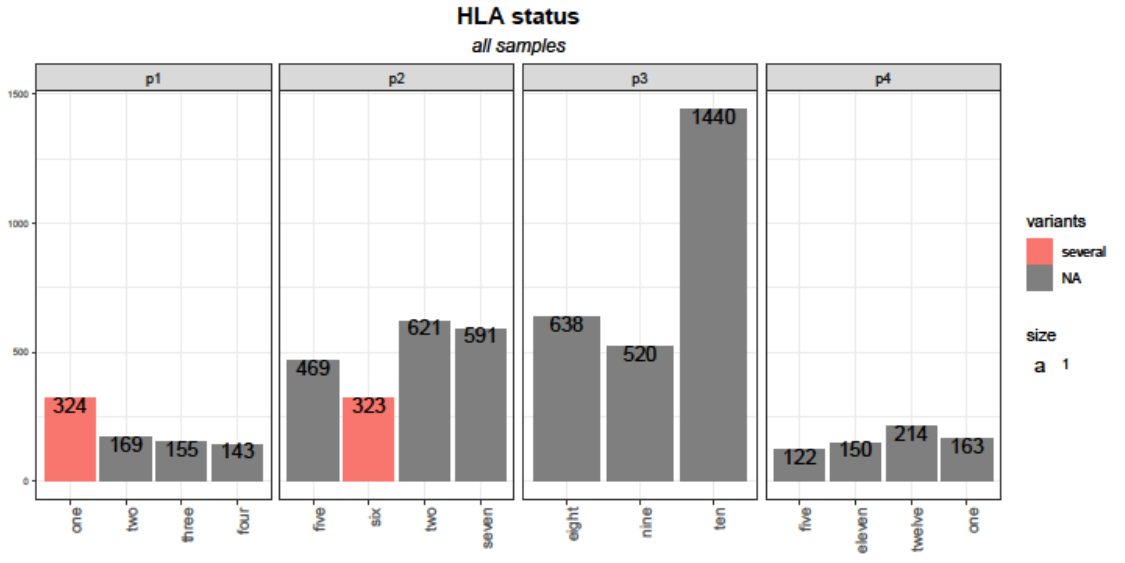
Instead I would suggest to map HLA on x, get rid of the scale_x_discrete and free your scales using scales="free_x".
If you want to reorder your bar chart I would suggest to set the factor levels of your HLA column accordingly. To this end I add a helper column HLA2 to your df to which I add the row number so that your two "one" categories get unique. Afterwards I use a function for the labels argument of scale_x_discrete to remove the row numbers:
library(ggplot2)
fig13_melt$HLA2 <- paste0(fig13_melt$HLA, seq(nrow(fig13_melt)))
fig13_melt$HLA2 <- forcats::fct_inorder(fig13_melt$HLA2)
ggplot(fig13_melt, aes(x = HLA2, y = value, fill = variants))
geom_bar(stat = "identity", position = "dodge")
geom_text(aes(label = round(value, 0), vjust = 1, size = 1), show.legend = FALSE)
labs(title = "HLA status", subtitle = "all samples", x = "HLA")
theme_bw()
theme(
axis.title.y = element_blank(), axis.title.x = element_blank(),
plot.title = element_text(face = "bold", size = 15, hjust = 0.5),
plot.subtitle = element_text(size = 12, hjust = 0.5, face = "italic"),
axis.text.y = element_text(size = 6),
axis.text.x = element_text(angle = 90, hjust = 1, size = 6)
)
scale_x_discrete(labels = ~ gsub("\\d $", "", .x))
facet_wrap(~NOI_ID, nrow = 1, scales = "free_x")
DATA
fig13_melt <- structure(list(NOI_ID = c(
"p1", "p1", "p1", "p1", "p2", "p2",
"p2", "p2", "p3", "p3", "p3", "p4", "p4", "p4", "p4"
), HLA = c(
"one",
"two", "three", "four", "five", "six", "two", "seven", "eight",
"nine", "ten", "five", "eleven", "twelve", "one"
), variants = c(
"several",
NA, NA, NA, NA, "several", NA, NA, NA, NA, NA, NA, NA, NA, NA
), variable = structure(c(
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L
), levels = "TPM", class = "factor"), value = c(
324.28,
169.21, 154.78, 143.31, 468.7, 322.76, 620.98, 591.17, 637.74,
519.8, 1439.58, 122.05, 149.74, 213.7, 162.53
)), row.names = c(
NA,
-15L
), class = "data.frame")
CodePudding user response:
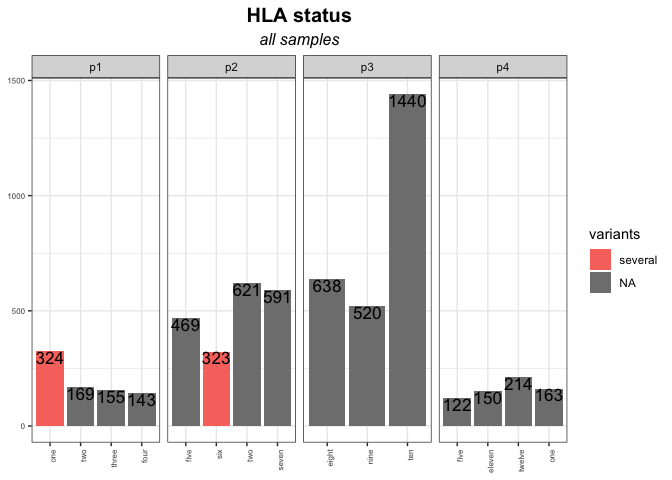
There are 3 parts to be revised:
- Use
x = factor(1:nrow(fig13_melt))inaes()to make the x-axis discrete instead of continuous. In this way, you can also change the label order of x-axis by settinglevels=...infactor().
ggplot(fig13_melt, aes(x = factor(1:nrow(fig13_melt)), y = value, fill = variants))
- Remove
limits = factor(1:nrow(fig13_melt))inscale_x_discrete(); Otherwise, all of the categories in the x-axis will be shown.
scale_x_discrete(labels = fig13_melt$HLA,
breaks = factor(1:nrow(fig13_melt)),
name='HLA')
- Add
scales = "free_x"intofacet_wrap():
facet_wrap(.~ NOI_ID, nrow = 1, scales = "free_x")
Data
fig13_melt <- structure(list(NOI_ID = c("p1", "p1", "p1", "p1", "p2", "p2",
"p2", "p2", "p3", "p3", "p3", "p4", "p4", "p4", "p4"), HLA = c("one",
"two", "three", "four", "five", "six", "two", "seven", "eight",
"nine", "ten", "five", "eleven", "twelve", "one"), variants = c("several",
NA, NA, NA, NA, "several", NA, NA, NA, NA, NA, NA, NA, NA, NA
), variable = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L), levels = "TPM", class = "factor"), value = c(324.28,
169.21, 154.78, 143.31, 468.7, 322.76, 620.98, 591.17, 637.74,
519.8, 1439.58, 122.05, 149.74, 213.7, 162.53)), row.names = c(NA,
-15L), class = "data.frame")