I need help because I'm not skilled enough to accomplish that task. I'm going to have several .csv files saved this way (with more and more rows):
ExamTitle ParametersName ParametersValue
1 Titolo nuovo esame TrunkLenght_VPDM_SAG 446
2 Titolo nuovo esame TrunkInclination_VPDM_SAG -0,49
3 Titolo nuovo esame SagittalImbalance_SAG -4
4 Titolo nuovo esame CervicalLordosisDepth_SAG 30
And I will export them all within a df with this code.
setwd("C:/Users/directory")
tbl_fread <- list.files(pattern = "*.csv") %>%
map_df(~fread(.))
However, I'd need your help because I want to move the terms in the column ParametersName to "separate columns". I give you my idea about how the df should look.
name1<-c("one", "two", "three", "four", "five")
name2<-c(1:5)
name3<-c(LETTERS[1:5])
name4<-c(letters[6:10])
df<-data.frame(name1, name2, name3,name4)
name1 name2 name3 name4
1 one 1 A f
2 two 2 B g
3 three 3 C h
4 four 4 D i
5 five 5 E j
The way I'd like to transform it by names3's values and drop names4 column ->
A B C D E
1 one two three four five
2 1 2 3 4 5
So once I have my df loaded with all the csv, I just pick the columns I need dividing by ParametersName's values.
Thank you guys.
CodePudding user response:
Try this:
## Loading the required libraries
library(dplyr)
library(tidyverse)
## Creating Dummy Data
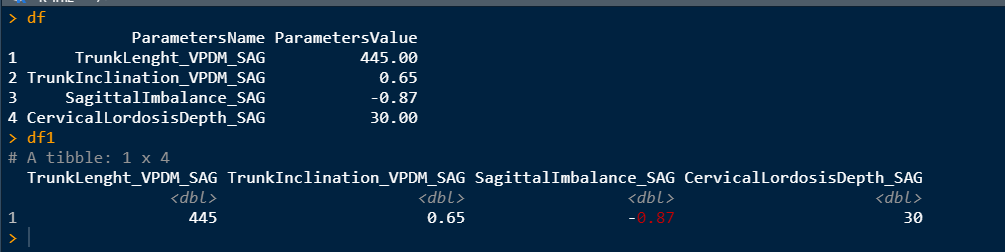
df = data.frame(ParametersName = c("TrunkLenght_VPDM_SAG","TrunkInclination_VPDM_SAG","SagittalImbalance_SAG","CervicalLordosisDepth_SAG"),
ParametersValue = c(445,0.65,-0.87,30))
df1 = df %>%
pivot_wider(names_from = ParametersName, values_from = ParametersValue)
CodePudding user response:
Solution with data.table
Since you're using data.table, here you can find a full data.table solution:
library(data.table)
# get files this way: it is preferable not to use setwd()
files <- list.files(dir, pattern = ".csv$", full.names = TRUE)
# fastest way to read your csv.
# drop here the column you don't want (I assumed it was ExamTitle)
# add id to reshape later
dt <- rbindlist(lapply(files, fread, drop = "ExamTitle"), idcol = "id")
# reshape with `data.table`
dcast(dt, id ~ ParametersName, value.var = "ParametersValue")
#> id CervicalLordosisDepth_SAG SagittalImbalance_SAG TrunkInclination_VPDM_SAG
#> 1: 1 30 -4 -0,49
#> 2: 2 30 -4 -0,49
#> TrunkLenght_VPDM_SAG
#> 1: 446
#> 2: 446
Solution with tidyverse
You can also use tidyverse. It depends on you and your project.
library(tidyverse)
# read and bind dataframes, add id
map_df(files, read_csv2, .id = "id") %>%
# remove column
select(-ExamTitle) %>%
# reshape
pivot_wider(names_from = ParametersName, values_from = ParametersValue)
#> # A tibble: 2 x 5
#> id TrunkLenght_VPDM_SAG TrunkInclination~ SagittalImbalan~ CervicalLordosi~
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 1 446 -0.49 -4 30
#> 2 2 446 -0.49 -4 30
Solution with Base R
And to conclude, you can also solve your problem with a one-line solution from base R
unstack(Reduce(rbind, lapply(files, read.csv2)), form = ParametersValue ~ ParametersName)
#> CervicalLordosisDepth_SAG SagittalImbalance_SAG TrunkInclination_VPDM_SAG TrunkLenght_VPDM_SAG
#> 1 30 -4 -0.49 446
#> 2 16 -4 -0.49 446
Reproducible example
Here, I'll leave a simple reproducible example to run my code.
dir <- tempdir()
write("ExamTitle;ParametersName;ParametersValue
Titolo nuovo esame;TrunkLenght_VPDM_SAG;446
Titolo nuovo esame;TrunkInclination_VPDM_SAG;-0,49
Titolo nuovo esame;SagittalImbalance_SAG;-4
Titolo nuovo esame;CervicalLordosisDepth_SAG;30",
file = file.path(dir, "tmp1.csv"))
write("ExamTitle;ParametersName;ParametersValue
Titolo nuovo esame;TrunkLenght_VPDM_SAG;446
Titolo nuovo esame;TrunkInclination_VPDM_SAG;-0,49
Titolo nuovo esame;SagittalImbalance_SAG;-4
Titolo nuovo esame;CervicalLordosisDepth_SAG;30",
file = file.path(dir, "tmp2.csv"))