I have a dataset that looks like this:
Study_ID time_point value
1 100 Time1 15
2 100 Time2 50
3 100 Time3 120
4 200 Time1 20
5 200 Time2 35
6 200 Time3 150
7 300 Time1 35
8 300 Time2 67
9 300 Time3 95
Where each patient (Study_ID) has 3 rows for the 3 time-points (Time 1, Time 2, and Time 3), with a value for each.
I would like to create a spaghetti plot with time_point on the x-axis, and the value on the y-axis, with a line for each patient. My desired output would look something like this:
How can I go about doing this?
Reproducible data:
data<-data.frame(Study_ID=c("100","100","100","200","200","200","300","300","300"),time_point=c("Time1","Time2","Time3","Time1","Time2","Time3","Time1","Time2","Time3"),value=c("15","50","120","20","35","150","35","67","95"))
CodePudding user response:
By using the group and color arguments within aes() you can then add the layers geom_point() and geom_line() to keep color and group together.
library(tidyverse)
data<-data.frame(Study_ID=c("100","100","100","200","200","200","300","300","300"),time_point=c("Time1","Time2","Time3","Time1","Time2","Time3","Time1","Time2","Time3"),value=c("15","50","120","20","35","150","35","67","95"))
ggplot(data, aes(time_point, value, group = Study_ID, color = Study_ID))
geom_point()
geom_line()
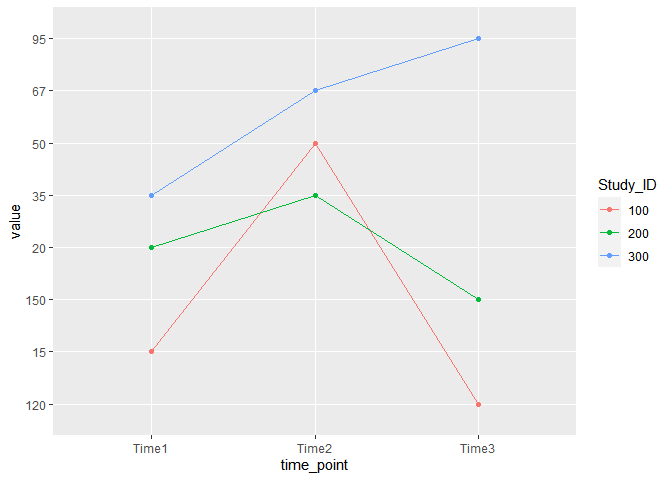
Created on 2022-06-20 by the reprex package (v2.0.1)