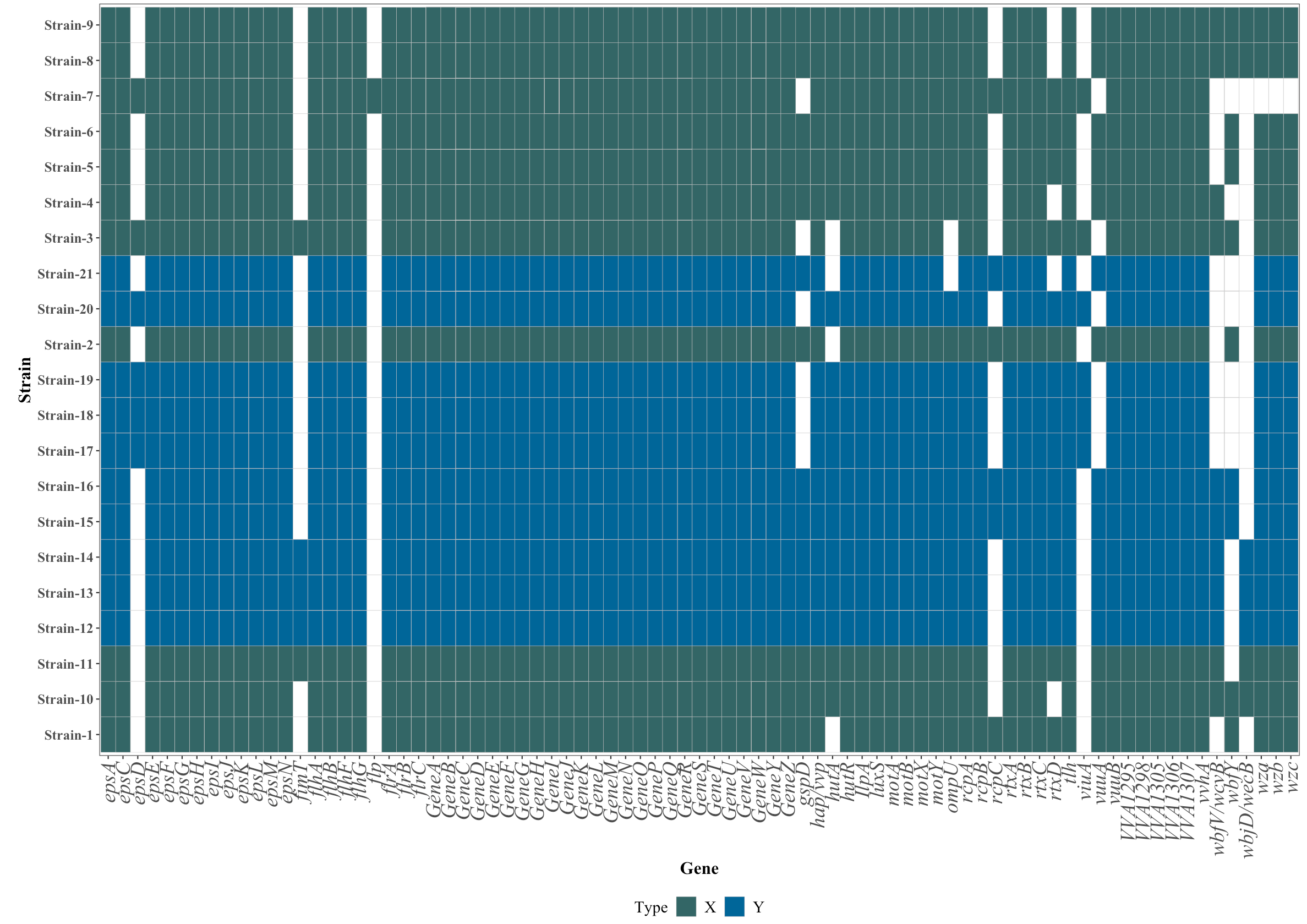
I have a data frame:
ID Strain value Type Gene
182 VFG007613(gi:27366370) Strain-1 0 X motY
183 VFG007614(gi:37679367) Strain-1 1 X motY
184 VFG007619(gi:27364700) Strain-1 0 X motX
185 VFG007620(gi:37681249) Strain-1 1 X motX
186 VFG007622(gi:27364235) Strain-1 0 X wza
187 VFG007623(gi:37678521) Strain-1 1 X wza
188 VFG007627(gi:37678523) Strain-1 1 X wzb
189 VFG007629(gi:27364230) Strain-1 1 X wzc
190 VFG007630(gi:37678524) Strain-1 0 X wzc
191 VFG007640(gi:37678525) Strain-1 0 X wbjD/wecB
192 VFG007653(gi:27364224) Strain-1 1 X wbfY
193 VFG007654(gi:37678548) Strain-1 0 X wbfY
194 VFG007656(gi:27364223) Strain-1 0 X wbfV/wcvB
195 VFG007657(gi:37678549) Strain-1 0 X wbfV/wcvB
196 VFG007668(gi:27367928) Strain-1 1 X GeneA
197 VFG007669(gi:37676055) Strain-1 0 X GeneA
The name of the genes are like wbfY, always the last letter are in upper case, so I want to plot the axis.text.x using the column Gene in italic except the upper case, something like: wbfY, wbjD/wecB, motY, motX, geneG, anynameA. It possible to use some function to plot all the text in italic( genename) except the upper case notitalic(:upper:): genenameD.
Or any way to do it ?
ggplot(mydf, aes(x=Gene, y=Strain, fill = Type, alpha = value))
scale_fill_manual(values = c("#336666", "#006699"))
geom_tile(colour = "grey79")
scale_alpha_identity(guide = "none")
coord_fixed(ratio = 2.4)
theme_bw()
theme(text = element_text(family = "Times New Roman"), # corresponde a todo en Time New Romans
legend.position="bottom",
legend.title = element_text(size = 14), # legend size title, corresponde a Genotype
legend.text = element_text(size = 14), # corresponde a vcgC and vcgE
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.text.x = element_text(angle = 90, size = 18, hjust =1, vjust = 0.5, face = "italic"),
axis.title.x = element_text(size = 15, face = "bold"),
axis.text.y = element_text(angle = 0, hjust = 1, size = 12, face = "bold"),
axis.title.y = element_text(size = 15, face = "bold")
)
in this case all axis.text.x are all in italic but I want to avoid the upper case !!!
How can I make that ?
Thanks so much
CodePudding user response:
What you can do is split the gene is parts and use glue to make the last letter italic using element_rmarkdown from ggtext like this:
library(tidyverse)
library(stringi)
library(glue)
library(ggtext)
mydf %>%
mutate(Gene_last_letter = stri_sub(Gene, -1, -1),
Gene_first_letters = str_sub(Gene, 1, str_length(Gene)-1)) %>%
mutate(Gene = glue("<i>{Gene_first_letters}</i>{Gene_last_letter}")) %>%
ggplot(aes(x=Gene, y=Strain, fill = Type, alpha = value))
scale_fill_manual(values = c("#336666", "#006699"))
geom_tile(colour = "grey79")
scale_alpha_identity(guide = "none")
coord_fixed(ratio = 2.4)
theme_bw()
theme(text = element_text(family = "Times New Roman"), # corresponde a todo en Time New Romans
legend.position="bottom",
legend.title = element_text(size = 14), # legend size title, corresponde a Genotype
legend.text = element_text(size = 14), # corresponde a vcgC and vcgE
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.text.x = element_markdown(angle = 90, size = 18, hjust =1, vjust = 0.5),
axis.title.x = element_text(size = 15, face = "bold"),
axis.text.y = element_text(angle = 0, hjust = 1, size = 12, face = "bold"),
axis.title.y = element_text(size = 15, face = "bold")
)
Output: