I'm trying to visualize a grouped boxplot with p-values in r.
I've tried plotting the box plot, and it works but while performing the t-test and then plotting the significant p-values, I'm facing an issue.
Instead of the numerical p-values, it should print a * for the significant values else ns.
My attempt has been:
ttcluster_dataset %>%
tidyr::pivot_longer(cols = -Phenotype) %>%
ggplot()
geom_boxplot(aes(
x =Phenotype,
y = value,
color=Phenotype,
fill= Phenotype))
labs(x = "Phenotypes",
y = "Gene Expression Value")
scale_fill_brewer(palette="RdBu") guides(fill=guide_legend(title="Phenotypes"),color=guide_legend(title="Phenotypes")) ggtitle("Cluster Gene Expression across Phenotypes")
The Dataset is:
ttcluster_dataset[,595:597]<-structure(list(ZNF8 = c(6.535976, 5.617546, 5.520076, 6.173408,
5.636716, 5.600262, 5.220575, 4.743357, 4.901029, 4.925283, 5.838014,
4.596382, 4.914675, 5.377216, 5.555028, 5.314098, 5.297864, 5.750704,
5.488065, 4.999103, 5.992091, 5.642773, 5.07518, 5.314475, 5.900937,
5.559608, 4.522685, 5.265154, 5.324593, 5.354034, 5.7289, 5.057386,
5.06977, 5.597334, 5.153986, 5.108475, 5.570181, 4.899142, 5.406266,
5.113253, 4.960403, 5.435193, 6.867495, 4.856415, 5.111619, 5.119095,
4.760636, 5.416521, 5.363732, 4.495227, 4.902833, 6.109712, 4.667864,
5.449276, 5.198133, 5.388347, 5.282403, 4.632593, 4.829716, 4.811047,
5.32258, 4.610439, 5.18732, 5.64755, 5.482928, 5.119356, 5.444986,
5.477327, 4.885934, 5.349004, 5.66351, 4.303683, 4.592721, 5.422557,
5.087755, 4.716865, 4.669027, 5.340175, 4.782326, 5.649563, 5.132957,
5.683762, 5.932028, 7.316127, 6.146593, 5.72388, 5.966438, 4.792467,
5.720676, 5.148644, 5.73848, 6.212466, 6.308748, 5.379114, 5.272329,
5.439909, 5.586977, 5.09346, 5.576321, 6.207303, 5.592295, 4.593619,
4.889057, 5.293568, 4.586061, 5.428235, 5.8397, 5.96753, 5.801161,
5.987631, 6.203965, 5.015435, 5.181961, 5.834061, 4.864606, 5.081433,
5.220504, 4.839773, 4.715386, 4.636048, 5.860811, 6.033741, 4.733141,
5.138581, 5.114714, 5.466906, 4.908799, 5.060534, 5.255573, 4.830971,
5.263964, 5.525848, 5.220203, 4.957148, 4.740335, 4.5796, 4.594491,
5.352114, 5.157587, 5.436527, 4.91852, 5.08691, 5.023482, 5.534429,
5.051983, 5.253279, 5.040294, 5.216557, 4.901129, 5.324232, 4.74494,
5.136233, 5.024926, 5.33796, 4.793476, 4.707207, 5.811963, 5.502805,
5.038211, 4.890697, 4.606382, 4.869951, 4.930496, 4.52962, 4.493444,
4.526999, 4.635502, 5.040377, 4.765426, 4.493035, 5.218385),
ZNF804A = c(6.51568, 6.439824, 4.177717, 5.722987, 7.93036,
8.825149, 6.117847, 5.202625, 6.943736, 6.10238, 5.395202,
4.005275, 5.449567, 5.250869, 5.375079, 6.279801, 4.846173,
4.312459, 4.968536, 7.3861, 8.261597, 7.374785, 6.239196,
5.368542, 5.101474, 5.054613, 5.31017, 4.846913, 5.216192,
5.737364, 5.338054, 4.207094, 6.386935, 4.983061, 7.574587,
4.413107, 5.352411, 6.507691, 5.199225, 5.900246, 5.338985,
4.046878, 8.342389, 4.816628, 5.722269, 4.192456, 4.246963,
6.287769, 4.228797, 4.226257, 4.474805, 4.336341, 4.362907,
4.635663, 4.8078, 4.223784, 5.340559, 5.434575, 4.493881,
4.84106, 6.921161, 4.690488, 6.626063, 4.473161, 4.489687,
4.323735, 4.360158, 4.583291, 6.180486, 4.410217, 6.529253,
4.332168, 5.635954, 4.46047, 5.444109, 4.409367, 4.607851,
4.415356, 3.9169, 4.028548, 4.286836, 4.498551, 5.981919,
4.136005, 4.24244, 4.089674, 4.29059, 3.859174, 4.062029,
4.326703, 4.236188, 4.27184, 3.95776, 4.491866, 4.209833,
4.202512, 4.158955, 4.174552, 5.370753, 4.933963, 3.982912,
4.162634, 4.101793, 4.332177, 4.263031, 5.336449, 4.02567,
4.168412, 4.131943, 4.263566, 4.107345, 4.182984, 4.173398,
4.151008, 4.37511, 4.066581, 4.050416, 4.111515, 4.241965,
4.483141, 4.170102, 4.146675, 4.479603, 4.473624, 4.667704,
4.019978, 4.190319, 4.442643, 4.456675, 5.083862, 4.649441,
4.57102, 4.110476, 4.423792, 4.461519, 4.309157, 3.784853,
4.249824, 4.127431, 4.136897, 4.779541, 4.261828, 4.150655,
4.605232, 4.489233, 4.284094, 5.285193, 4.413362, 4.528692,
4.108301, 5.526067, 5.096504, 4.480856, 4.271599, 5.242742,
4.213557, 4.74649, 4.027772, 4.640365, 4.344877, 4.077101,
4.283787, 4.534188, 4.24613, 4.056275, 4.55178, 4.29397,
5.215219, 4.295077, 4.434866, 5.129624), Phenotype = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L, 8L, 8L, 8L,
8L, 8L, 8L, 8L, 8L), .Label = c("Proneural (Cluster 1)",
"Proneural (Cluster 2)", "Neural (Cluster 1)", "Neural (Cluster 2)",
"Classical (Cluster 1)", "Classical (Cluster 2)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 2)"), class = "factor")), class = "data.frame", row.names = c("TCGA.02.0014.01",
"TCGA.02.0026.01", "TCGA.02.0048.01", "TCGA.02.0069.01", "TCGA.02.0074.01",
"TCGA.02.0080.01", "TCGA.02.0084.01", "TCGA.02.0087.01", "TCGA.02.0104.01",
"TCGA.02.0114.01", "TCGA.02.0281.01", "TCGA.02.0321.01", "TCGA.02.0325.01",
"TCGA.02.0338.01", "TCGA.02.0339.01", "TCGA.02.0432.01", "TCGA.02.0439.01",
"TCGA.02.0440.01", "TCGA.02.0446.01", "TCGA.06.0128.01", "TCGA.06.0129.01",
"TCGA.06.0146.01", "TCGA.06.0156.01", "TCGA.06.0166.01", "TCGA.06.0174.01",
"TCGA.06.0177.01", "TCGA.06.0238.01", "TCGA.06.0241.01", "TCGA.06.0410.01",
"TCGA.06.0413.01", "TCGA.06.0414.01", "TCGA.06.0646.01", "TCGA.06.0648.01",
"TCGA.08.0245.01", "TCGA.08.0344.01", "TCGA.08.0347.01", "TCGA.08.0348.01",
"TCGA.08.0350.01", "TCGA.08.0353.01", "TCGA.08.0359.01", "TCGA.08.0385.01",
"TCGA.08.0517.01", "TCGA.08.0524.01", "TCGA.12.0616.01", "TCGA.12.0618.01",
"TCGA.02.0003.01", "TCGA.02.0010.01", "TCGA.02.0011.01", "TCGA.02.0024.01",
"TCGA.02.0028.01", "TCGA.02.0046.01", "TCGA.02.0047.01", "TCGA.02.0060.01",
"TCGA.02.0089.01", "TCGA.02.0113.01", "TCGA.02.0115.01", "TCGA.02.0451.01",
"TCGA.06.0132.01", "TCGA.06.0133.01", "TCGA.06.0138.01", "TCGA.06.0160.01",
"TCGA.06.0162.01", "TCGA.06.0167.01", "TCGA.06.0171.01", "TCGA.06.0173.01",
"TCGA.06.0179.01", "TCGA.06.0182.01", "TCGA.06.0185.01", "TCGA.06.0195.01",
"TCGA.06.0208.01", "TCGA.06.0214.01", "TCGA.06.0219.01", "TCGA.06.0221.01",
"TCGA.06.0237.01", "TCGA.06.0240.01", "TCGA.08.0349.01", "TCGA.08.0380.01",
"TCGA.08.0386.01", "TCGA.08.0520.01", "TCGA.02.0016.01", "TCGA.02.0023.01",
"TCGA.02.0070.01", "TCGA.02.0102.01", "TCGA.02.0260.01", "TCGA.02.0269.01",
"TCGA.02.0285.01", "TCGA.02.0289.01", "TCGA.02.0290.01", "TCGA.02.0317.01",
"TCGA.02.0333.01", "TCGA.02.0422.01", "TCGA.02.0430.01", "TCGA.06.0125.01",
"TCGA.06.0126.01", "TCGA.06.0137.01", "TCGA.06.0145.01", "TCGA.06.0148.01",
"TCGA.06.0187.01", "TCGA.06.0211.01", "TCGA.06.0402.01", "TCGA.08.0246.01",
"TCGA.08.0354.01", "TCGA.08.0355.01", "TCGA.08.0357.01", "TCGA.08.0358.01",
"TCGA.08.0375.01", "TCGA.08.0511.01", "TCGA.08.0514.01", "TCGA.08.0518.01",
"TCGA.08.0529.01", "TCGA.08.0531.01", "TCGA.02.0007.01", "TCGA.02.0009.01",
"TCGA.02.0021.01", "TCGA.02.0027.01", "TCGA.02.0038.01", "TCGA.02.0043.01",
"TCGA.02.0004.01", "TCGA.02.0039.01", "TCGA.02.0059.01", "TCGA.02.0064.01",
"TCGA.02.0075.01", "TCGA.02.0079.01", "TCGA.02.0085.01", "TCGA.02.0086.01",
"TCGA.02.0099.01", "TCGA.02.0106.01", "TCGA.02.0107.01", "TCGA.02.0111.01",
"TCGA.02.0326.01", "TCGA.02.0337.01", "TCGA.06.0122.01", "TCGA.06.0124.01",
"TCGA.06.0143.01", "TCGA.06.0147.01", "TCGA.06.0149.01", "TCGA.06.0152.01",
"TCGA.06.0154.01", "TCGA.06.0164.01", "TCGA.06.0175.01", "TCGA.06.0176.01",
"TCGA.06.0184.01", "TCGA.06.0189.01", "TCGA.06.0190.01", "TCGA.06.0194.01",
"TCGA.06.0197.01", "TCGA.06.0210.01", "TCGA.06.0397.01", "TCGA.06.0409.01",
"TCGA.06.0412.01", "TCGA.06.0644.01", "TCGA.06.0645.01", "TCGA.08.0346.01",
"TCGA.08.0352.01", "TCGA.08.0360.01", "TCGA.08.0390.01", "TCGA.08.0509.01",
"TCGA.08.0510.01", "TCGA.08.0512.01", "TCGA.12.0619.01", "TCGA.12.0620.01",
"TCGA.02.0025.01", "TCGA.02.0033.01", "TCGA.02.0034.01", "TCGA.02.0051.01",
"TCGA.02.0054.01", "TCGA.02.0057.01", "TCGA.06.0130.01", "TCGA.06.0139.01",
"TCGA.08.0392.01", "TCGA.08.0522.01"))
All suggestions will be welcomed.
CodePudding user response:
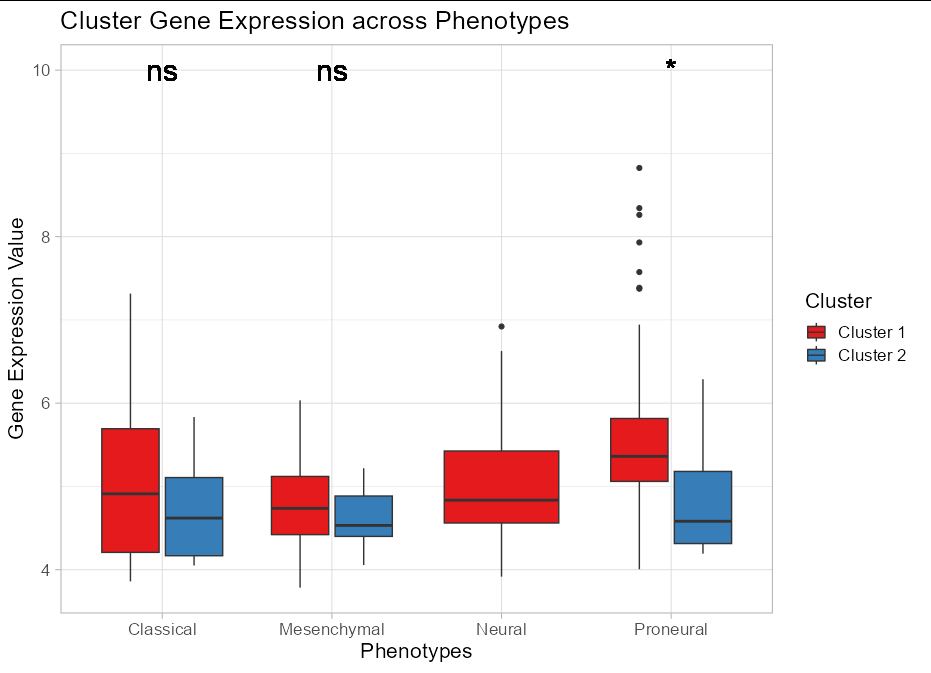
I'm guessing that you want the p value between clusters for each phenotype. I would probably start by tidying your data so that Phenotype and Cluster are different variables, each with their own column, since logically they are distinct. This will aid calculation of the p values and help with plotting.
Secondly, from a data visualization point of view, having each phenotype / cluster having a different fill color doesn't really convey any information. You should use fill color to denote either cluster or phenotype. Filling by the interaction of the two simply makes it harder to interpret what the colors mean. Also, the x axis is already labelled with the same values ad the fill legend, so the fill colors are really just a bit gratuitous. Using a completely unrelated color scale for the outlines makes the whole thing look a bit garish.
Fixing these issues, I might present the data something like this:
ttcluster_dataset %>%
tidyr::pivot_longer(cols = -Phenotype) %>%
separate(Phenotype, into = c("Phenotype", "Cluster"), sep = " \\(") %>%
mutate(Cluster = sub("\\)", "", Cluster)) %>%
group_by(Phenotype) %>%
mutate(pval = ifelse(first(Phenotype == "Neural"), 1,
t.test(value[Cluster == "Cluster 1"],
value[Cluster == "Cluster 2"])$p.val)) %>%
mutate(pval = ifelse(pval == 1, "", ifelse(pval < 0.05, "*", "ns"))) %>%
ggplot()
geom_boxplot(aes(
x =Phenotype,
y = value,
fill = Cluster))
geom_text(aes(x = Phenotype, y = 10, label = pval), size = 8)
labs(x = "Phenotypes",
y = "Gene Expression Value")
scale_fill_brewer(palette = "Set1")
theme_light(base_size = 16)
ggtitle("Cluster Gene Expression across Phenotypes")
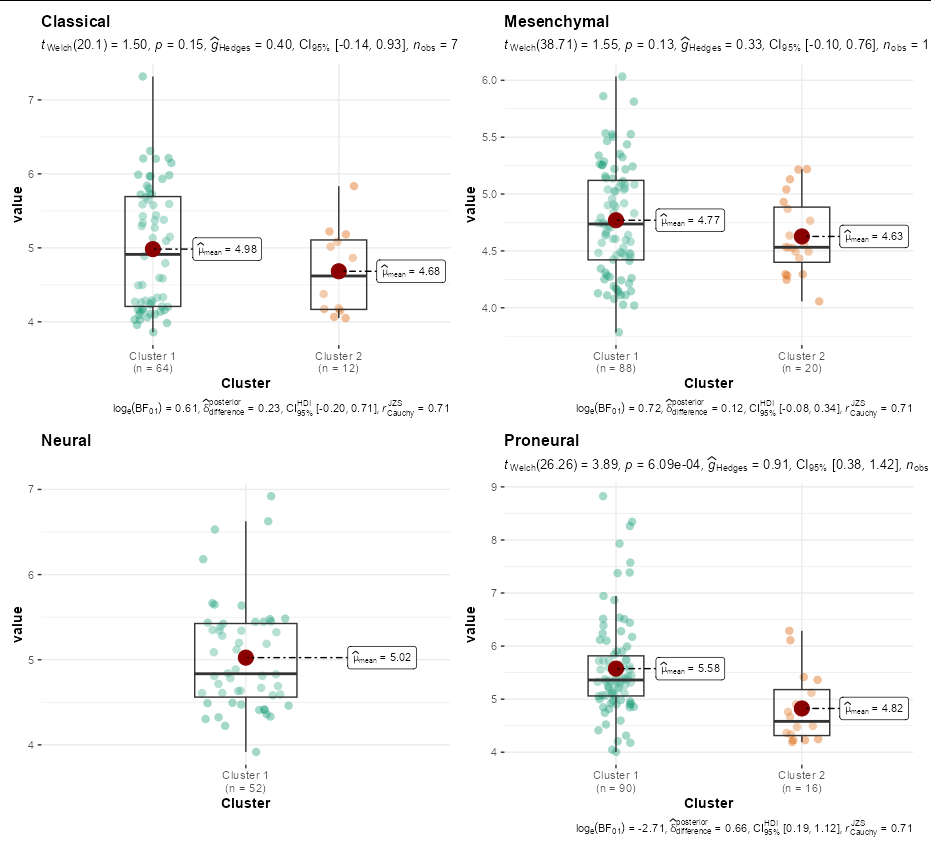
You might even consider a more sophisticated plot using grouped_ggbetweenstats from ggstatsplot
ttcluster_dataset %>%
tidyr::pivot_longer(cols = -Phenotype) %>%
separate(Phenotype, into = c("Phenotype", "Cluster"), sep = " \\(") %>%
mutate(Cluster = sub("\\)", "", Cluster)) %>%
group_by(Phenotype) %>%
ggstatsplot::grouped_ggbetweenstats(x = Cluster, y = "value",
plot.type = "box",
grouping.var = Phenotype)