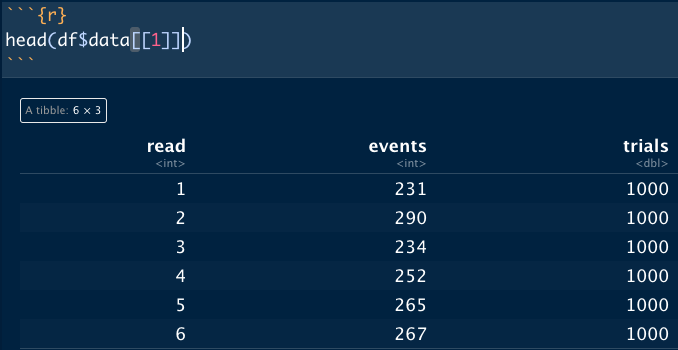
I want to generate 1 million simulated datasets and save them in a list column.
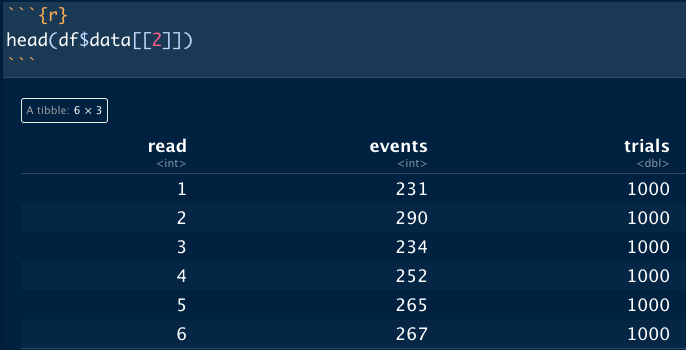
The problem I run into is that every generated dataset is identical, which I assume is a result of the operation being vectorized. Is there a way to maintain the speed of a vectorized operation while changing the seed for each execution of rbinom()?
library(tidyverse)
SIMS <- 100
df <-
tibble(
sim = 1:5,
data =
list(
tibble(
read = 1:100,
events = rbinom(n=SIMS, size=1e3, prob = .25),
trials = 1e3
)
)
)
df
head(df$data[[1]])
head(df$data[[2]])
I have tried using parallel::mclapply(), which solves the seed issue, but the performance is nowhere near as fast as the vectorized operation.
BASERATE <- .25
DAILY_N <- 1e3
DURATION <- 500
time_to_insight <- function(){
tibble(
read = 1:DURATION,
events = rbinom(n=DURATION, size=DAILY_N, prob = BASERATE),
trials = DAILY_N
)
}
temp <-
parallel::mclapply(
X = 1:1000,
FUN = time_to_insight,
mc.cores = 8L
)
CodePudding user response:
Depending on how your simulation works, it might be possible to create all the data for all the simulations first, and then group them into a list to get the original format with unique data.
n = 5
SIMS = 100
set.seed(42)
df <-
tibble(
sim = rep(1:n, each = SIMS),
read = rep(1:SIMS, times = n),
events = rbinom(n=SIMS*n, size=1e3, prob = .25),
trials = 1e3) %>%
nest(data = -sim)
Result
> head(df$data[[1]])
# A tibble: 6 × 3
read events trials
<int> <int> <dbl>
1 1 240 1000
2 2 262 1000
3 3 240 1000
4 4 250 1000
5 5 214 1000
6 6 244 1000
> head(df$data[[2]])
# A tibble: 6 × 3
read events trials
<int> <int> <dbl>
1 1 247 1000
2 2 260 1000
3 3 253 1000
4 4 245 1000
5 5 259 1000
6 6 247 1000
CodePudding user response:
You can (almost) maintain the speed of your original function by generating the realisations of your random variable (events) by generating all of the realisations for all of your simulations at the same time. This is possible because your event probability remains constant for all the simulations.
Here's how I would do it. First write a function to generate all your simulations at the same time.
generateTibble <- function() {
tibble() %>%
expand(sim=1:5, read=1:100, trials=1000) %>%
mutate(events=rbinom(n=nrow(.), size=1000, prob=0.25))
}
And use it
df <- generateTibble()
df
# A tibble: 500 × 4
sim read trials events
<int> <int> <dbl> <int>
1 1 1 1000 246
2 1 2 1000 232
3 1 3 1000 258
4 1 4 1000 233
5 1 5 1000 259
6 1 6 1000 251
7 1 7 1000 254
8 1 8 1000 263
9 1 9 1000 267
10 1 10 1000 266
# … with 490 more rows
Demonstrate that realisations do not repeat across sims:
df %>% group_by(sim) %>% group_map(head, n=2)
[[1]]
# A tibble: 2 × 3
read trials events
<int> <dbl> <int>
1 1 1000 246
2 2 1000 232
[[2]]
# A tibble: 2 × 3
read trials events
<int> <dbl> <int>
1 1 1000 255
2 2 1000 261
[[3]]
# A tibble: 2 × 3
read trials events
<int> <dbl> <int>
1 1 1000 234
2 2 1000 239
[[4]]
# A tibble: 2 × 3
read trials events
<int> <dbl> <int>
1 1 1000 252
2 2 1000 258
[[5]]
# A tibble: 2 × 3
read trials events
<int> <dbl> <int>
1 1 1000 255
2 2 1000 258
Compare the speed of the new function with the original (faulty) version:
SIMS <- 100
originalFunction <- function() {
df <-
tibble(
sim = 1:5,
data =
list(
tibble(
read = 1:100,
events = rbinom(n=SIMS, size=1e3, prob = .25),
trials = 1e3
)
)
)
df
}
library(microbenchmark)
microbenchmark(originalFunction, generateTibble, times=100)
Unit: nanoseconds
expr min lq mean median uq max neval
originalFunction 42 43 52.95 44 44 877 100
generateTibble 47 48 68.14 48 54 1403 100
So, a 28% increase in mean execution time. Is that reasonable?