I have two dataframes. One has demographics information about patients and other has some feature information. Below is some dummy data representing my dataset:
Demographics:
demographics = {
'PatientID': [10, 11, 12, 13],
'DOB': ['1971-10-23', '1969-06-18', '1973-04-20', '1971-05-31'],
'Sex': ['M', 'M', 'F', 'M'],
'Flag': [0, 1, 0, 0]
}
demographics = pd.DataFrame(demographics)
demographics['DOB'] = pd.to_datetime(demographics['DOB'])
Here is the printed dataframe:
print(demographics)
PatientID DOB Sex Flag
0 10 1971-10-23 M 0
1 11 1969-06-18 M 1
2 12 1973-04-20 F 0
3 13 1971-05-31 M 0
Features:
features = {
'PatientID': [10, 10, 10, 10, 10, 10, 10, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12],
'Feature': ['A', 'B', 'A', 'A', 'C', 'B', 'C', 'A', 'B', 'B', 'A', 'C', 'D', 'A', 'B', 'C', 'C', 'D', 'D', 'D', 'B', 'C', 'C', 'C', 'B', 'B', 'C'],
}
features = pd.DataFrame(features)
Here is a count of each features of each patient:
print(features.groupby(['PatientID', 'Feature']).size())
PatientID Feature
10 A 3
B 2
C 2
11 A 3
B 3
C 3
D 1
12 B 3
C 4
D 3
dtype: int64
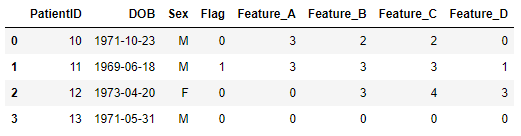
I want to integrate each patients feature counts of their features into the demographics table. Note that patient 13 is absent from the features table. The final dataframe will look as shown below:
result = {
'PatientID': [10, 11, 12, 13],
'DOB': ['1971-10-23', '1969-06-18', '1973-04-20', '1971-05-31'],
'Feature_A': [3, 3, 0, 0],
'Feature_B': [2, 3, 3, 0],
'Feature_C': [2, 3, 4, 0],
'Feature_D': [0, 1, 3, 0],
'Sex': ['M', 'M', 'F', 'M'],
'Flag': [0, 1, 0, 0],
}
result = pd.DataFrame(result)
result['DOB'] = pd.to_datetime(result['DOB'])
print(result)
PatientID DOB Feature_A Feature_B Feature_C Feature_D Sex Flag
0 10 1971-10-23 3 2 2 0 M 0
1 11 1969-06-18 3 3 3 1 M 1
2 12 1973-04-20 0 3 4 3 F 0
3 13 1971-05-31 0 0 0 0 M 0
How can I get this result from these two dataframes?
CodePudding user response:
Cross-tabulate features and merge with demographics.
# cross-tabulate feature df
# and reindex it by PatientID to carry PatientIDs without features
feature_counts = (
pd.crosstab(features['PatientID'], features['Feature'])
.add_prefix('Feature_')
.reindex(demographics['PatientID'], fill_value=0)
)
# merge the two
demographics.merge(feature_counts, on='PatientID')
CodePudding user response:
Fix your code adding unstack
out = (features.groupby(['PatientID', 'Feature']).size().
unstack(fill_value=0).
add_prefix('Feature_').
reindex(demographics['PatientID'],fill_value=0).
reset_index().
merge(demographics))
Out[30]:
PatientID Feature_A Feature_B Feature_C Feature_D DOB Sex Flag
0 10 3 2 2 0 1971-10-23 M 0
1 11 3 3 3 1 1969-06-18 M 1
2 12 0 3 4 3 1973-04-20 F 0
3 13 0 0 0 0 1971-05-31 M 0