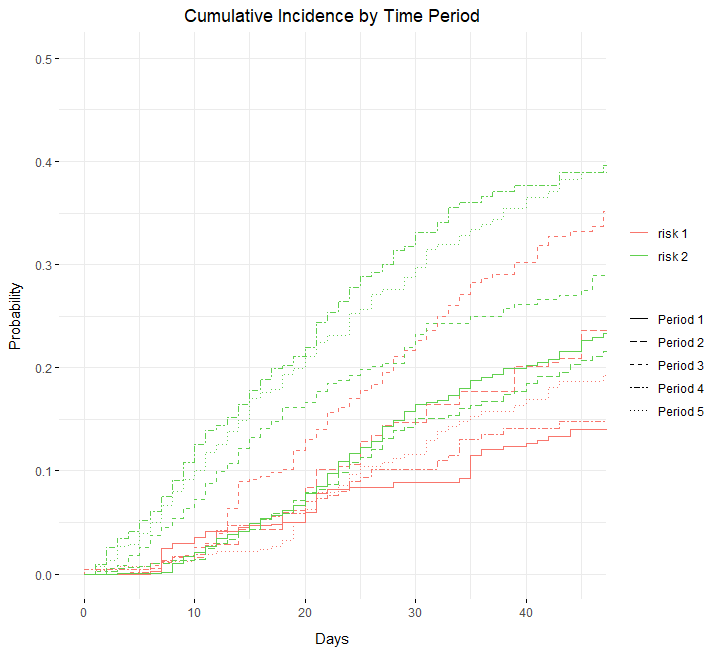
I created a plot using ggcompetingrisks that is illegible. I would like to color code the time periods and differentiate the risk groups by using dashed vs dotted lines. Right now it's flipped.
p3 <- ggcompetingrisks(fit = TimePer_fit,
multiple_panels = F,
ggtheme = theme_minimal(),
coef = 1.96, #95% CI
conf.int = F,
xlim = c(0, 45),
ylim = c(0, 0.5),
xlab = "Days",
ylab = "Probability",
title = "Cumulative Incidence by Time Period")
theme(plot.title = element_text(hjust = 0.5), #center title
axis.title.y = element_text(margin = margin(t = 0, r = 10, b = 0, l = 0)), #pad y axis label
axis.title.x = element_text(margin = margin(t = 10, r = 0, b = 0, l = 0))) #pad x axis label
scale_color_manual(labels = c("risk 1", "risk 2"),
values = c("#F8766D", "619CFF"),
name = element_blank()) # do not name legend
scale_linetype_manual(labels = c("Period 1","Period 2","Period 3","Period 4", "Period 5"),
values = c("solid", "longdash", "dashed","twodash", "dotted"),
name = element_blank())
CodePudding user response:
Perhaps what you could try is to choose two contrasting colors. Also you can use this function facet_wrap(. ~ x), using as parameter either risk or period, and in this way you can have a clean plot.
CodePudding user response:
We can look into the plot object and see that colour is automatically mapped to event and linetype is mapped to group. These are not easily swapped using normal methods, as they are in the quosure form. (The quosure: a data structure that captures an expression along with its associated environment, as found in function arguments. -Hadley). But we can use other methods.
Here is a dummy example from the survminer package.
library(ggplot2)
library(cmprsk)
library(survminer)
set.seed(2)
ss <- rexp(100)
gg <- factor(sample(1:3,100,replace=TRUE),1:3,c('BRCA','LUNG','OV'))
cc <- factor(sample(0:2,100,replace=TRUE),0:2,c('no event', 'death', 'progression'))
strt <- sample(1:2,100,replace=TRUE)
# handles cuminc objects
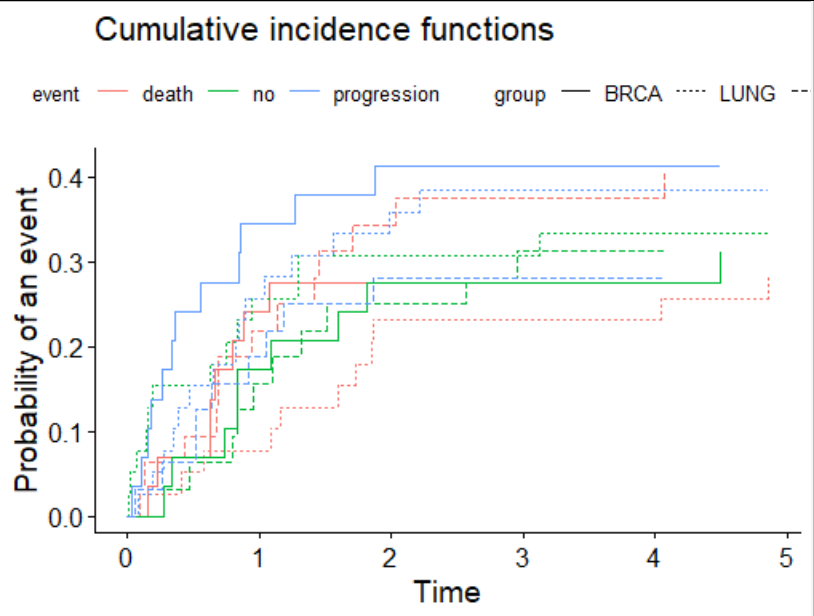
print(fit <- cmprsk::cuminc(ss,cc,gg,strt))
g<-ggcompetingrisks(fit,
multiple_panels = F)
g
We can see event is mapped to colour, and group to linetype.
g$mapping$colour <quosure> expr: ^event env: 0x00000295351a3b58
g$mapping$linetype <quosure> expr: ^group env: 0x00000295351a3b58
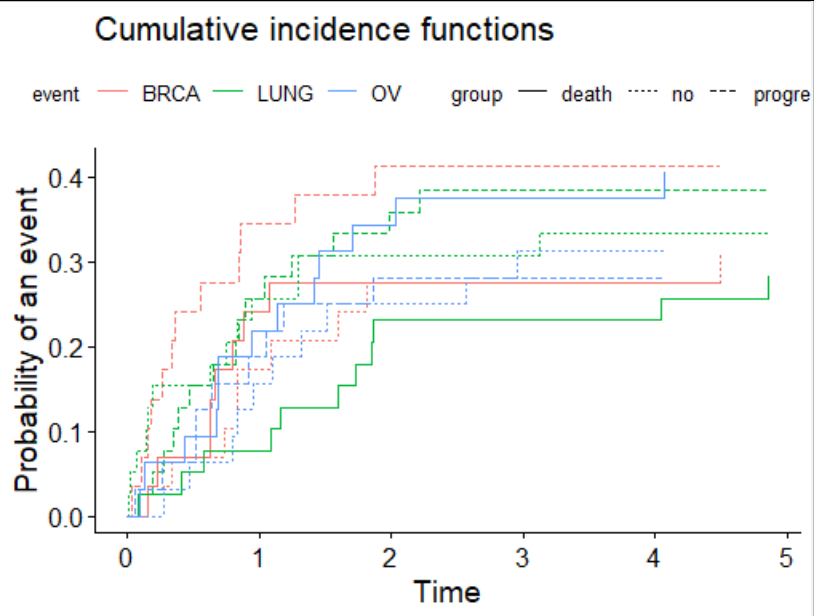
Now we access those elements in the plot object and swap them over.
library(rlang)
g$mapping$colour<-quo_set_expr(g$mapping$colour, quote(group))
g$mapping$linetype<-quo_set_expr(g$mapping$linetype, quote(event))
g