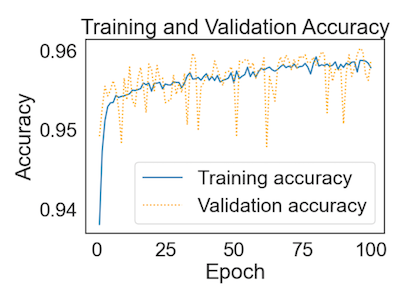
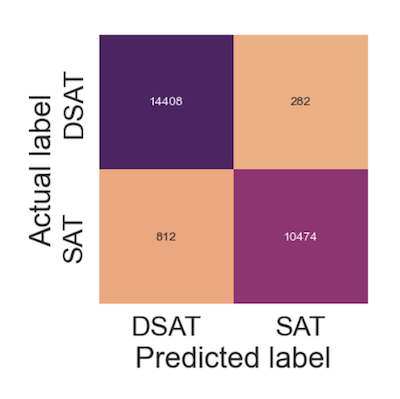
I'm performing binary classification on a Kaggle dataset that is known to be quite balanced and it has a shape of (129880,22).
My precision on the negative class is very low and it's zero on the positive class. roc-auc = 0.9934% and the true positives and true negatives are in line with the data.
Since I'm performing binary classification with 22 inputs, do I have too many Dense layers? I experimented manually and with BayesSearchCV, but not very familiar with the api.
def get_model():
return keras.Sequential([
keras.layers.Dense(22, input_dim=22, activation=tf.nn.relu, name = "Dense_1"),
keras.layers.Dense(22, activation=tf.nn.relu, name = "Dense_2"),
keras.layers.Dense(11, activation=tf.nn.relu, name = "Dense_3"),
keras.layers.Dense(1, activation=tf.nn.sigmoid, name = "sigmoid")
])
keras_model = get_model()
keras_model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=0.01),
loss=tf.keras.losses.binary_crossentropy,
metrics=["accuracy"]
)
keras_model.summary()
keras_model.fit(scaled_X_train, y_train, steps_per_epoch=5, epochs=100, callbacks = earlystopping)
keras_model.evaluate(scaled_X_test, y_test, batch_size=10)
y_pred = (keras_model.predict(scaled_X_test) > 0.5)
y_predict_label = np.argmax(y_pred, axis=1)
report = classification_report(y_test.flatten(), y_pred.flatten())
precision recall f1-score support
0 0.57 1.00 0.72 14690
1 0.00 0.00 0.00 11286
accuracy 0.57 25976
macro avg 0.28 0.50 0.36 25976
weighted avg 0.32 0.57 0.41 25976
CodePudding user response:
the reason why you are getting low precision in the classification_report is because y_pred is the correct input to classification_report instead of y_predict_label (because this is a binary classification problem), so the last line should look as follows.
report = classification_report(y_test.flatten(), y_pred.flatten())
as for the validation plot, it is not uncommon for it to be noisy, this is because of the 'noise' in the training process (which includes quantization noise due to training on a subset of the data).
CodePudding user response:
Thanks to @ahmed-aek for giving me a clue to flatten y_test because of the dimensions mismatch.
I had to add values in the line y_test.values.flatten() because flatten is a method of np.array not of pd.Series.
Now the classification report looks copacetic.
precision recall f1-score support
0 0.95 0.98 0.96 14690
1 0.97 0.93 0.95 11286
accuracy 0.96 25976
macro avg 0.96 0.95 0.96 25976
weighted avg 0.96 0.96 0.96 25976