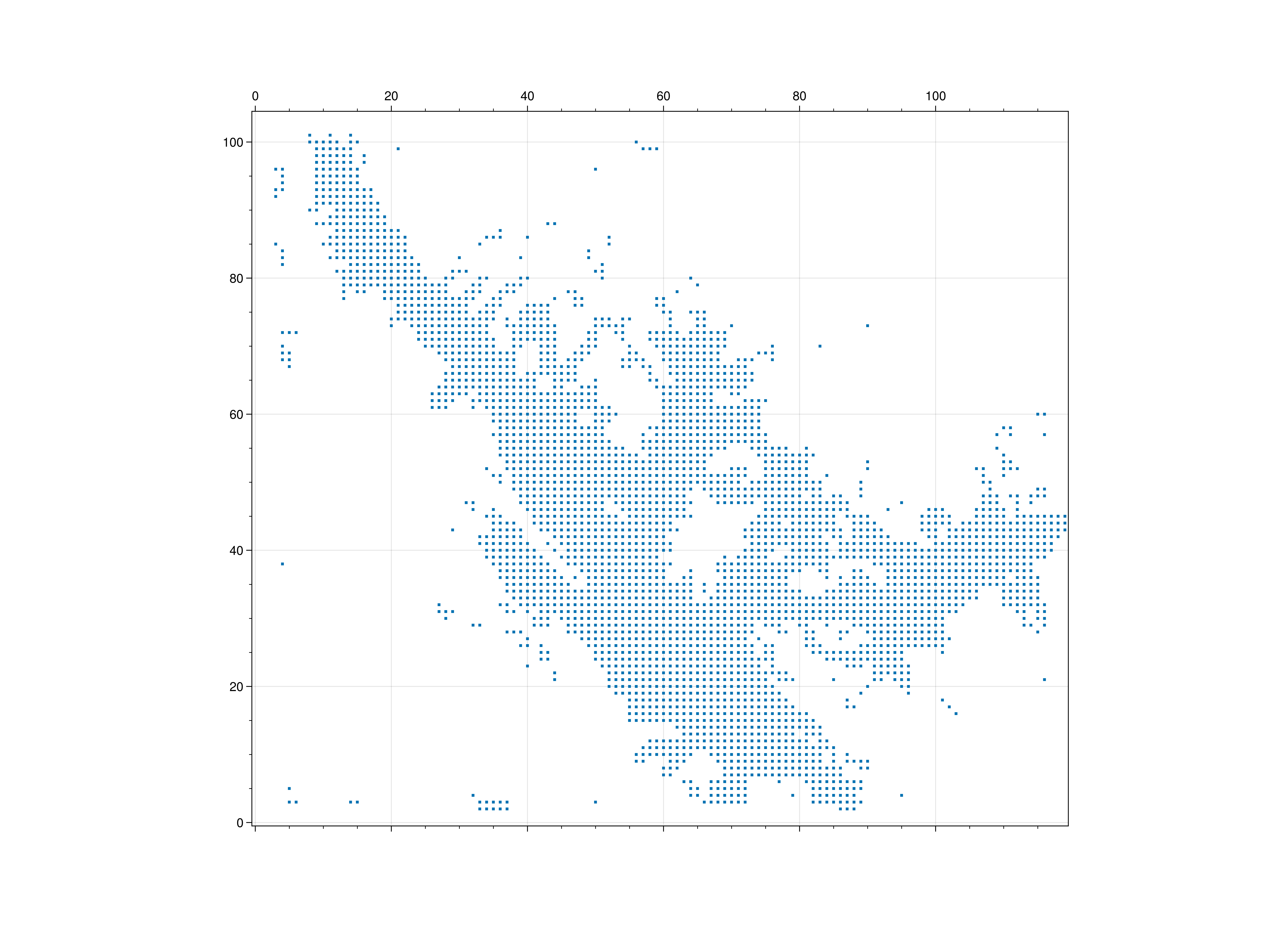
I have two sets of gridded (NetCDF) data with dimensions: Time, S-N, W-E and would like to do a t-test of paired values in time across the entire grid with Python. The computation should return decisions across the entire grid that can then be processed into a spy plot aerial overlay. I am able to process this in MATLAB, after reading the variable, comprising two main steps (as a hint to what is expected in Python):
**[h,p] = ttest2(permute(grid_1,[3 2 1]),permute(griid_2,[3 2 1]))** % [3 2 1] ===> [Time S-N W-E]
**spy(squeeze(h),'k ',1)** % h is 1/0 decision.
Perhaps this would entail array transforms but my knowledge of Python is limited.
A snippet of my gridded data is as follows:
<xarray.DataArray 'WSPD' (Time: 120, south_north: 105, west_east: 120)>
array([[[3.0042849, 3.6635756, 3.5766048, ..., 2.7890186, 3.5537026,
2.4510043],
Coordinates:
XLONG (south_north, west_east) float32 -125.7 -125.7 ... -122.3 -122.2
XLAT (south_north, west_east) float32 55.7 55.7 55.7 ... 56.99 56.99
XTIME (Time) float32 3.003e 04 7.282e 04 ... 1.417e 06 3.742e 06
Time (Time) datetime64[ns] 2010-08-01T08:00:00 ... 2020-07-01T08:00:00
CodePudding user response:
You can perform paired t-test using 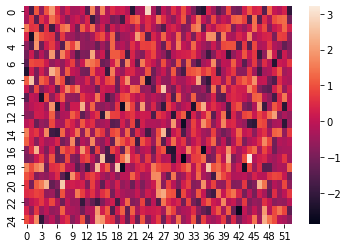
For plotting some decision, e.g. when pvalue is lower than 0.05, we can simply do
sns.heatmap(res.pvalue < 0.05, cbar=True) # Turn off the color bar
CodePudding user response:
What a relief!! Thank you so much for the simplicity of your solution! What I want for the final outcome follows from the following lines:
[statres,pval] = ttest_rel(grid_1, grid_2) # returns statistics, and the p-value
spy1 = np.where(pval < 0.05, 1, 0) # 1/0 array
ax = plt.gca()
ax.spy(spy1, precision=0.1, markersize=1)
ax.invert_yaxis() # to get the right direction
plt.show()
With this, I get finely dotted plots. How can I overlay on the gridded netcdf area? I mean, the x,y co-ordinates of the present plot are grid point #s, but I would like that it is mapped to the lat-lon coordinates of the netcdf grid. Also, the markersize arg changes the points to blue, but I would prefer black dots.