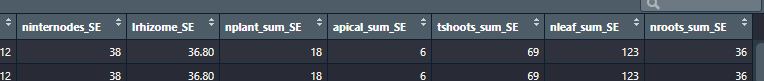
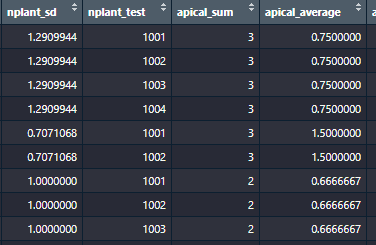
Thanks to @julian answer, I can manage this by:
sheet <- sheet %>%
group_by(across(all_of(.GlobalEnv$filter_list[[select_filter]]))) %>%
summarise(
across(
where(is.numeric),
list(sum = sum, average = mean, sd = sd),
.names = "{.col}_{.fn}"
)
)
Although this brings out another problem, where if I define my own function, the data is calculated on each value instead of by grouping:
testfunc <- function(.) {
. 1000
}
sheet <- sheet %>%
group_by(across(all_of(.GlobalEnv$filter_list[[select_filter]]))) %>%
summarise(
across(
where(is.numeric),
list(sum = sum, average = mean, sd = sd, test = testfunc),
.names = "{.col}_{.fn}"
#total = sum(.data[[names(sheet[,10])]], na.rm = TRUE)
)
)
CodePudding user response:
Use a list of functions inside the mutate
sheet <- sheet %>%
group_by(across(all_of(.GlobalEnv$filter_list[[select_filter]]))) %>%
mutate(
across(where(is.numeric), list(sum = sum, average = mean, sd = sd) , .names="{.col}_{.fn}")
)
But it's better to use summarise instead of mutate
sheet <- sheet %>%
group_by(across(all_of(.GlobalEnv$filter_list[[select_filter]]))) %>%
summarise(
across(where(is.numeric), list(sum = sum, average = mean, sd = sd) , .names="{.col}_{.fn}")
)
Example with theiris dataset
iris %>%
mutate(
across(where(is.numeric), list(sum = sum, average = mean, sd = sd) , .names="{.col}_{.fn}"))