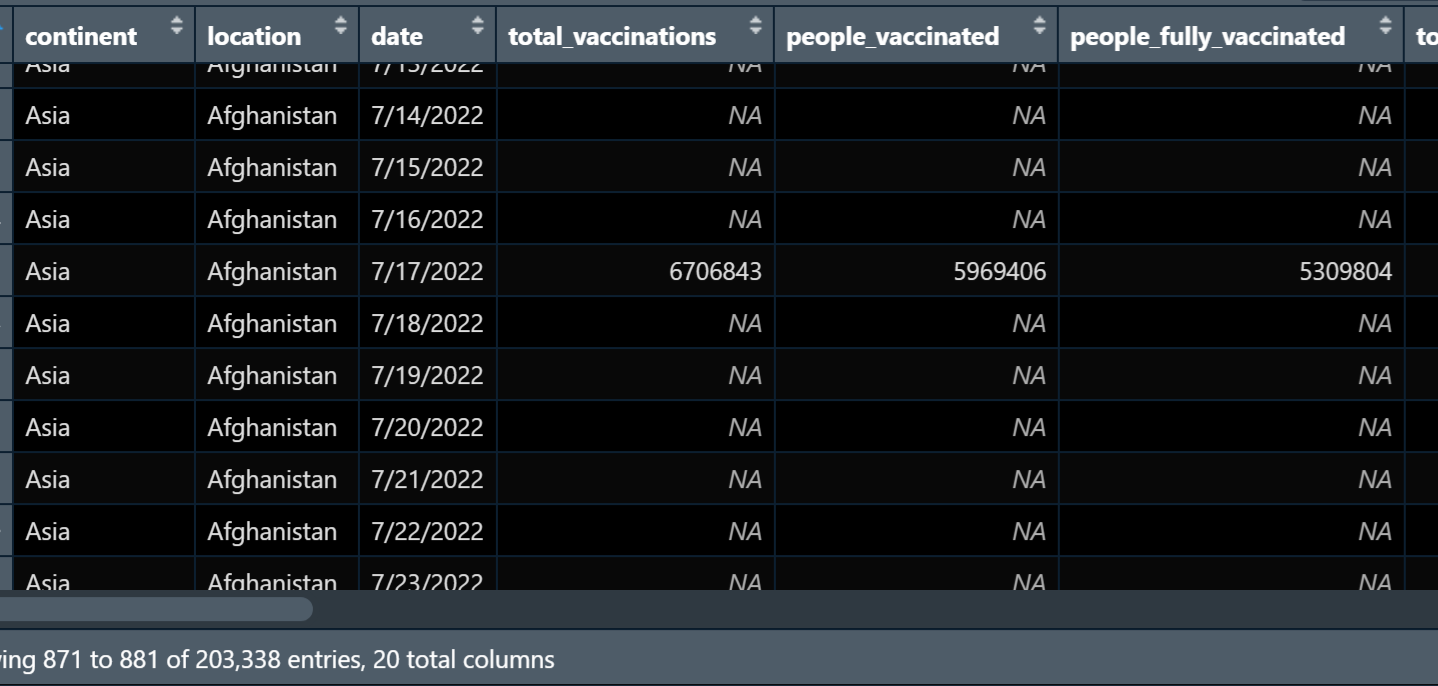
I'm trying to aggregate this df by the last value in each corresponding country observation. For some reason, the last value that is added to the tibble is not correct.
aggre_data <- combined %>%
group_by(location) %>%
summarise(Last_value_vacc = last(people_vaccinated_per_hundred)
aggre_data
I believe it has something to do with all of the NA values throughout the df. However I did try:
aggre_data <- combined %>%
group_by(location) %>%
summarise(Last_value_vacc = last(people_vaccinated_per_hundred(na.rm = TRUE)))
aggre_data
CodePudding user response:
combined %>%
group_by(location) %>%
arrange(date) %>% # or whatever
summarise(Last_value_vacc = last(people_vaccinated_per_hundred, na.rm = TRUE))
CodePudding user response:
If this is your dataset:
structure(list(continent = c("Asia", "Asia", "Asia", "Asia",
"Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia",
"Asia", "Asia", "Asia", "Asia"), location = c("China", "China",
"China", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan",
"Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan",
"Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan"),
date = c("7/16/2022", "7/17/2022", "7/18/2022", "7/19/2022",
"7/20/2022", "7/21/2022", "7/22/2022", "7/14/2022", "7/15/2022",
"7/16/2022", "7/17/2022", "7/18/2022", "7/19/2022", "7/20/2022",
"7/21/2022", "7/22/2022"), total_vaccinations = c(NA, 6706843L,
NA, NA, NA, NA, NA, NA, NA, NA, 6706843L, NA, NA, NA, NA,
1L), people_vaccinated = c(NA, 5969406L, NA, NA, NA, NA,
NA, NA, NA, NA, 5969406L, NA, NA, NA, NA, 2L), people_fully_vaccinated = c(NA,
5309804L, NA, NA, NA, NA, NA, NA, NA, NA, 5309804L, NA, NA,
NA, NA, 3L)), class = "data.frame", row.names = c(NA, -16L
))
continent location date total_vaccinations people_vaccinated people_fully_vaccinated
1 Asia China 7/16/2022 NA NA NA
2 Asia China 7/17/2022 6706843 5969406 5309804
3 Asia China 7/18/2022 NA NA NA
4 Asia Afghanistan 7/19/2022 NA NA NA
5 Asia Afghanistan 7/20/2022 NA NA NA
6 Asia Afghanistan 7/21/2022 NA NA NA
7 Asia Afghanistan 7/22/2022 NA NA NA
8 Asia Afghanistan 7/14/2022 NA NA NA
9 Asia Afghanistan 7/15/2022 NA NA NA
10 Asia Afghanistan 7/16/2022 NA NA NA
11 Asia Afghanistan 7/17/2022 6706843 5969406 5309804
12 Asia Afghanistan 7/18/2022 NA NA NA
13 Asia Afghanistan 7/19/2022 NA NA NA
14 Asia Afghanistan 7/20/2022 NA NA NA
15 Asia Afghanistan 7/21/2022 NA NA NA
16 Asia Afghanistan 7/22/2022 1 2 3
and you apply this code:
library(dplyr)
df %>%
group_by(location) %>%
arrange(date, .by_group = TRUE) %>%
summarise(Last_value_vacc = last(people_vaccinated))
then you get this
location Last_value_vacc
<chr> <int>
1 Afghanistan 2
2 China NA
