Below is an example of my dataset:
structure(list(wheezing_InDMod = c(0, 0, 0, 0, 0, 0, 0, 0, 1,
1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0), cough_anyMod = c(0,
0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0), SOB_anyMod = c(0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0,
1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), country.x = c("cameroon",
"cameroon", "cameroon", "kenya", "cameroon", "ghana", "cameroon",
"kenya", "cameroon", "kenya", "cameroon", "cameroon", "cameroon",
"cameroon", "cameroon", "cameroon", "cameroon", "cameroon", "ghana",
"cameroon", "kenya", "cameroon", "ghana", "cameroon", "cameroon",
"cameroon")), row.names = 65:90, class = "data.frame")
For wheezing_InDMod, SOB_anyMod & cough_anyMod, 1 indicates that this individual has the symptom and 0 indicates they do not
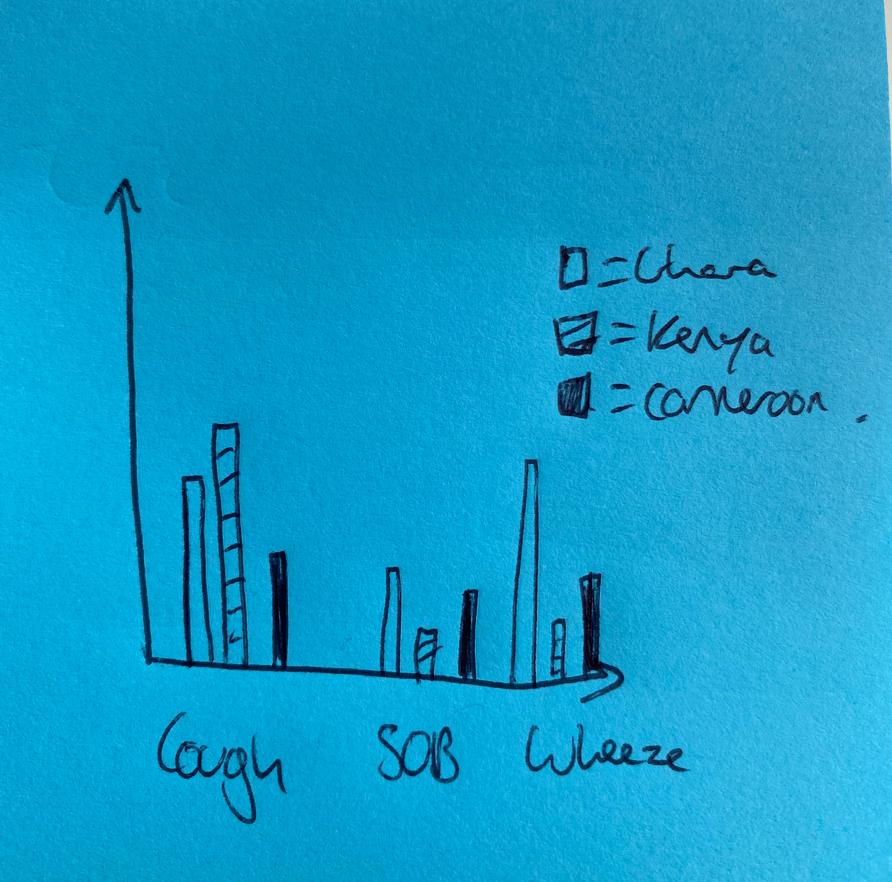
I'm trying to plot a single bargraph showing the prevalence of each symptom [ e.g. "1" for wheezing_InDMod, SOB_anyMod & cough_anyMod] on the X axis, with each further split into 3 adjacent bars based on the country.x category. I'll attach an image below to show an idea of what I mean:
Does anyone know how I would go about creating this using ggplot? I've tried a few different codes and I haven't got very far
CodePudding user response:
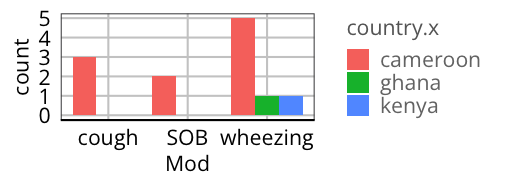
The single column thing isn't an issue. Here's an example using the code from my comment. I'd be curious to see the code that you tried that made you think this was an issue.
library(dplyr)
library(tidyr)
library(ggplot2)
your_data %>%
tidyr::pivot_longer(contains("Mod"), names_to = "Mod") %>%
## keep only 1s
filter(value == 1) %>%
## clean up the names
mutate(Mod = stringr::str_remove(Mod, "_.*")) %>%
ggplot(aes(x = Mod, fill = country.x))
geom_bar(position = position_dodge(preserve = "single"))