I'm a beginner of Python coding.
I have an exercise that use numpy to solve this Iris_df excercise.
Id sepal_length sepal_width petal_length petal_width species
0 1 5.1 3.5 1.4 0.2 Iris-setosa
1 2 4.9 3.0 1.4 0.2 Iris-setosa
2 3 4.7 3.2 1.3 0.2 Iris-setosa
3 4 4.6 3.1 1.5 0.2 Iris-setosa
4 5 5.0 3.6 1.4 0.2 Iris-setosa
... ... ... ... ... ... ...
145 146 6.7 3.0 5.2 2.3 Iris-virginica
146 147 6.3 2.5 5.0 1.9 Iris-virginica
147 148 6.5 3.0 5.2 2.0 Iris-virginica
148 149 6.2 3.4 5.4 2.3 Iris-virginica
149 150 5.9 3.0 5.1 1.8 Iris-virginica
150 rows × 6 columns
i don't know how to write the function to calculate min, max of each iris feature (sepal_length, sepal_width, petal_length, petal_width) but take each species separately (Iris-versicolor, Iris-setosa, Iris-virginica) (each species is 50 row)
Anyone can help me with this.
CodePudding user response:
Have you tried to use the groupby() function by pandas? With this function you can group ;) a dataframe based on a given key. On top of this group you can then apply pandas min(), max() etc.
If you provide some sample data I will edit my answer with an example.
CodePudding user response:
Get to feel about groupby() like function.
e.g., groupby().sum(),groupby().size(),groupby().min(), etc.
Then you're all good to use this kind of tools in everywhere.
df_min = df.groupby('species').min()
df_max = df.groupby('species').max()
df_min
###
sepal_length sepal_width petal_length petal_width
species
setosa 4.3 2.3 1.0 0.1
versicolor 4.9 2.0 3.0 1.0
virginica 4.9 2.2 4.5 1.4
df_max
###
sepal_length sepal_width petal_length petal_width
species
setosa 5.8 4.4 1.9 0.6
versicolor 7.0 3.4 5.1 1.8
virginica 7.9 3.8 6.9 2.5
via pivot_table()
(Metrics calculate the same things)
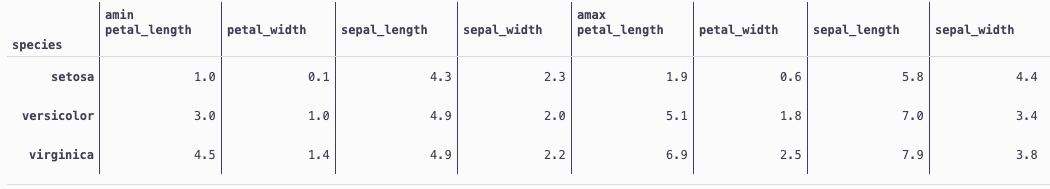
df.pivot_table(index='species', values=df.columns[:4],aggfunc=[np.min, np.max])
via groupby()
(Metrics could calculate different statistics)
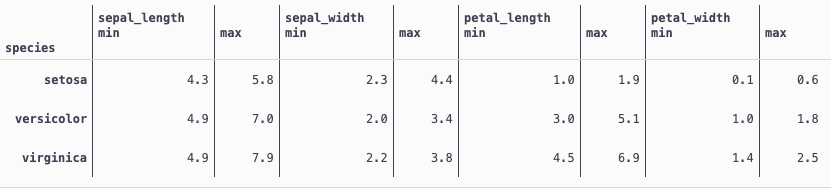
df.groupby('species').agg({'sepal_length':['min','max'],'sepal_width':['min','max'],'petal_length':['min','max'],'petal_width':['min','max']})