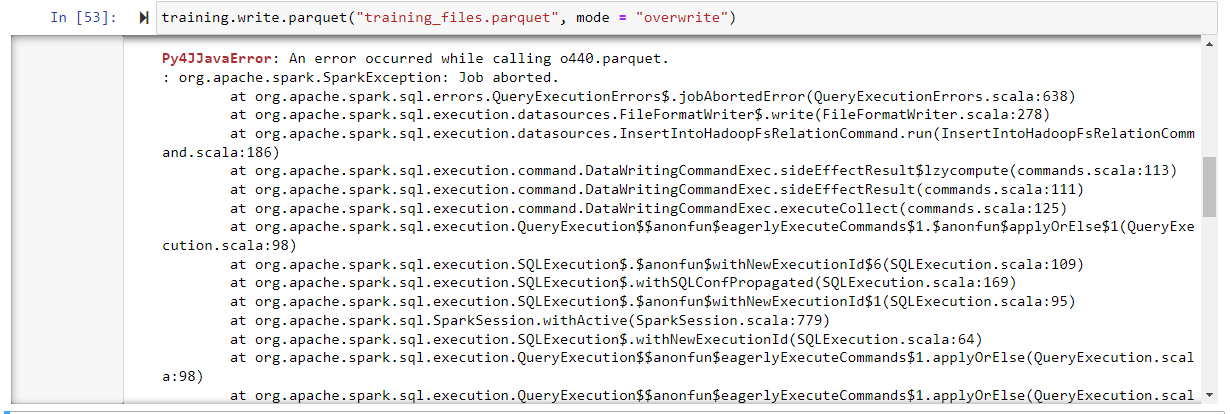
I'm trying to save the PySpark dataframe after transforming it using ML Pipeline. But when I save it the weird error is triggered every time.
Here are the columns of this dataframe:

And the following error occurs when I try to write the dataframe into parquet file format:
I tried to use different available winutils for Hadoop from 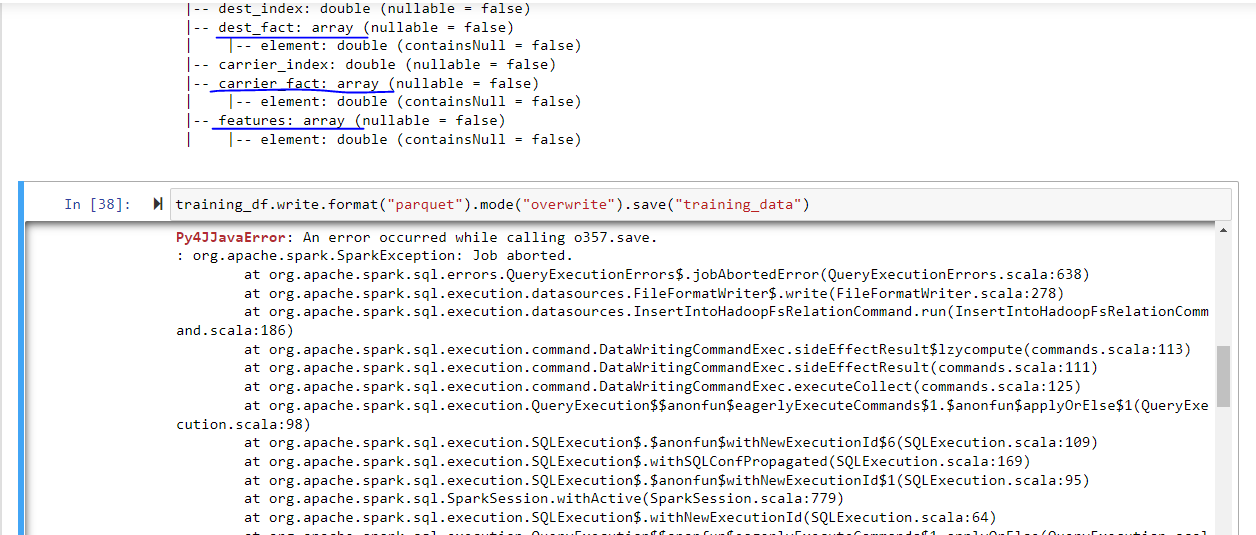
Thanks
Complete Error Message can be seen here:
---------------------------------------------------------------------------
Py4JJavaError Traceback (most recent call last)
~\AppData\Local\Temp\ipykernel_4448\2574092106.py in <cell line: 1>()
----> 1 training_df.write.format("parquet").mode("overwrite").save("training_data")
~\AppData\Local\Programs\Python\Python310\lib\site-packages\pyspark\sql\readwriter.py in save(self, path, format, mode, partitionBy, **options)
966 self._jwrite.save()
967 else:
--> 968 self._jwrite.save(path)
969
970 @since(1.4)
~\AppData\Local\Programs\Python\Python310\lib\site-packages\py4j\java_gateway.py in __call__(self, *args)
1319
1320 answer = self.gateway_client.send_command(command)
-> 1321 return_value = get_return_value(
1322 answer, self.gateway_client, self.target_id, self.name)
1323
~\AppData\Local\Programs\Python\Python310\lib\site-packages\pyspark\sql\utils.py in deco(*a, **kw)
188 def deco(*a: Any, **kw: Any) -> Any:
189 try:
--> 190 return f(*a, **kw)
191 except Py4JJavaError as e:
192 converted = convert_exception(e.java_exception)
~\AppData\Local\Programs\Python\Python310\lib\site-packages\py4j\protocol.py in get_return_value(answer, gateway_client, target_id, name)
324 value = OUTPUT_CONVERTER[type](answer[2:], gateway_client)
325 if answer[1] == REFERENCE_TYPE:
--> 326 raise Py4JJavaError(
327 "An error occurred while calling {0}{1}{2}.\n".
328 format(target_id, ".", name), value)
Py4JJavaError: An error occurred while calling o357.save.
: org.apache.spark.SparkException: Job aborted.
at org.apache.spark.sql.errors.QueryExecutionErrors$.jobAbortedError(QueryExecutionErrors.scala:638)
at org.apache.spark.sql.execution.datasources.FileFormatWriter$.write(FileFormatWriter.scala:278)
at org.apache.spark.sql.execution.datasources.InsertIntoHadoopFsRelationCommand.run(InsertIntoHadoopFsRelationCommand.scala:186)
at org.apache.spark.sql.execution.command.DataWritingCommandExec.sideEffectResult$lzycompute(commands.scala:113)
at org.apache.spark.sql.execution.command.DataWritingCommandExec.sideEffectResult(commands.scala:111)
at org.apache.spark.sql.execution.command.DataWritingCommandExec.executeCollect(commands.scala:125)
at org.apache.spark.sql.execution.QueryExecution$$anonfun$eagerlyExecuteCommands$1.$anonfun$applyOrElse$1(QueryExecution.scala:98)
at org.apache.spark.sql.execution.SQLExecution$.$anonfun$withNewExecutionId$6(SQLExecution.scala:109)
at org.apache.spark.sql.execution.SQLExecution$.withSQLConfPropagated(SQLExecution.scala:169)
at org.apache.spark.sql.execution.SQLExecution$.$anonfun$withNewExecutionId$1(SQLExecution.scala:95)
at org.apache.spark.sql.SparkSession.withActive(SparkSession.scala:779)
at org.apache.spark.sql.execution.SQLExecution$.withNewExecutionId(SQLExecution.scala:64)
at org.apache.spark.sql.execution.QueryExecution$$anonfun$eagerlyExecuteCommands$1.applyOrElse(QueryExecution.scala:98)
at org.apache.spark.sql.execution.QueryExecution$$anonfun$eagerlyExecuteCommands$1.applyOrElse(QueryExecution.scala:94)
at org.apache.spark.sql.catalyst.trees.TreeNode.$anonfun$transformDownWithPruning$1(TreeNode.scala:584)
at org.apache.spark.sql.catalyst.trees.CurrentOrigin$.withOrigin(TreeNode.scala:176)
at org.apache.spark.sql.catalyst.trees.TreeNode.transformDownWithPruning(TreeNode.scala:584)
at org.apache.spark.sql.catalyst.plans.logical.LogicalPlan.org$apache$spark$sql$catalyst$plans$logical$AnalysisHelper$$super$transformDownWithPruning(LogicalPlan.scala:30)
at org.apache.spark.sql.catalyst.plans.logical.AnalysisHelper.transformDownWithPruning(AnalysisHelper.scala:267)
at org.apache.spark.sql.catalyst.plans.logical.AnalysisHelper.transformDownWithPruning$(AnalysisHelper.scala:263)
at org.apache.spark.sql.catalyst.plans.logical.LogicalPlan.transformDownWithPruning(LogicalPlan.scala:30)
at org.apache.spark.sql.catalyst.plans.logical.LogicalPlan.transformDownWithPruning(LogicalPlan.scala:30)
at org.apache.spark.sql.catalyst.trees.TreeNode.transformDown(TreeNode.scala:560)
at org.apache.spark.sql.execution.QueryExecution.eagerlyExecuteCommands(QueryExecution.scala:94)
at org.apache.spark.sql.execution.QueryExecution.commandExecuted$lzycompute(QueryExecution.scala:81)
at org.apache.spark.sql.execution.QueryExecution.commandExecuted(QueryExecution.scala:79)
at org.apache.spark.sql.execution.QueryExecution.assertCommandExecuted(QueryExecution.scala:116)
at org.apache.spark.sql.DataFrameWriter.runCommand(DataFrameWriter.scala:860)
at org.apache.spark.sql.DataFrameWriter.saveToV1Source(DataFrameWriter.scala:390)
at org.apache.spark.sql.DataFrameWriter.saveInternal(DataFrameWriter.scala:363)
at org.apache.spark.sql.DataFrameWriter.save(DataFrameWriter.scala:239)
at java.base/jdk.internal.reflect.DirectMethodHandleAccessor.invoke(DirectMethodHandleAccessor.java:104)
at java.base/java.lang.reflect.Method.invoke(Method.java:577)
at py4j.reflection.MethodInvoker.invoke(MethodInvoker.java:244)
at py4j.reflection.ReflectionEngine.invoke(ReflectionEngine.java:357)
at py4j.Gateway.invoke(Gateway.java:282)
at py4j.commands.AbstractCommand.invokeMethod(AbstractCommand.java:132)
at py4j.commands.CallCommand.execute(CallCommand.java:79)
at py4j.ClientServerConnection.waitForCommands(ClientServerConnection.java:182)
at py4j.ClientServerConnection.run(ClientServerConnection.java:106)
at java.base/java.lang.Thread.run(Thread.java:833)
Caused by: java.lang.UnsatisfiedLinkError: 'boolean org.apache.hadoop.io.nativeio.NativeIO$Windows.access0(java.lang.String, int)'
at org.apache.hadoop.io.nativeio.NativeIO$Windows.access0(Native Method)
at org.apache.hadoop.io.nativeio.NativeIO$Windows.access(NativeIO.java:793)
at org.apache.hadoop.fs.FileUtil.canRead(FileUtil.java:1218)
at org.apache.hadoop.fs.FileUtil.list(FileUtil.java:1423)
at org.apache.hadoop.fs.RawLocalFileSystem.listStatus(RawLocalFileSystem.java:601)
at org.apache.hadoop.fs.FileSystem.listStatus(FileSystem.java:1972)
at org.apache.hadoop.fs.FileSystem.listStatus(FileSystem.java:2014)
at org.apache.hadoop.fs.ChecksumFileSystem.listStatus(ChecksumFileSystem.java:761)
at org.apache.hadoop.fs.FileSystem.listStatus(FileSystem.java:1972)
at org.apache.hadoop.fs.FileSystem.listStatus(FileSystem.java:2014)
at org.apache.hadoop.mapreduce.lib.output.FileOutputCommitter.getAllCommittedTaskPaths(FileOutputCommitter.java:334)
at org.apache.hadoop.mapreduce.lib.output.FileOutputCommitter.commitJobInternal(FileOutputCommitter.java:404)
at org.apache.hadoop.mapreduce.lib.output.FileOutputCommitter.commitJob(FileOutputCommitter.java:377)
at org.apache.parquet.hadoop.ParquetOutputCommitter.commitJob(ParquetOutputCommitter.java:48)
at org.apache.spark.internal.io.HadoopMapReduceCommitProtocol.commitJob(HadoopMapReduceCommitProtocol.scala:192)
at org.apache.spark.sql.execution.datasources.FileFormatWriter$.$anonfun$write$25(FileFormatWriter.scala:267)
at scala.runtime.java8.JFunction0$mcV$sp.apply(JFunction0$mcV$sp.java:23)
at org.apache.spark.util.Utils$.timeTakenMs(Utils.scala:642)
at org.apache.spark.sql.execution.datasources.FileFormatWriter$.write(FileFormatWriter.scala:267)
... 39 more
CodePudding user response:
For spark version 3.0.0 and up you can make use of pyspark.ml.functions.vector_to_array to convert the vector types to array types and then write.
from pyspark.ml.functions import vector_to_array
df = df.withColumn("dest_fact", vector_to_array("dest_fact"))\
.withColumn("features", vector_to_array("features"))
df.write.format("parquet").mode("overwrite").save("/file/output/path")
From the full stack trace, it seems there is also an issue with your hadoop setup. You could try the following steps as outlined in this post:
- Ensure you have set up
$HADOOP_HOMEenvironment variable - Ensure
%HADOOP_HOME%\binis added to thePATH - Download the appropriate
winutils.exeandhadoop.dllfor your hadoop version. You can try to find it in this github repo. - Place
winutils.exeandhadoop.dllinside thehadoop/binfolder. - Place
hadoop.dllinsideC:\Windows\System32.
