Problem:
Iterating over each row of a dataframe, find the row where the 'Datetime' value is less than or equal to the current 'Datetime' value - 20 seconds.
For example: if the previous 'Datetime' (at index i-1) is at least 20s earlier than the current one - it will be chosen. If not (e.g. only 5 seconds earlier), move to i-2 and see if it is at least 20s earlier. Until the condition is met.
NaT in the beginning since there is no time before it.
When the previous 'Datetime' is found - it will be put into a new column, and also new columns will be appended for the other values in this row (row with the previous 'Datetime').
Example result:
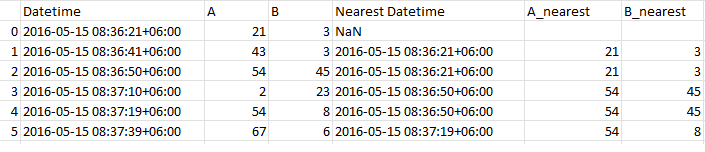
The last three columns are the newly created columns, and the first three columns are the original dataset.
CodePudding user response:
Edit: 100x faster way using np.searchsorted
Using np.searchsorted to calculate z below makes this 100x faster: 3.31 ms for 10K rows.
import numpy as np
def select_2(df, min_dt='20s'):
newcols = [f'{k}_nearest' for k in df.columns]
s = df['Datetime']
z = np.searchsorted(s, s - (pd.Timedelta(min_dt) - pd.Timedelta('1ns'))) - 1
return pd.concat([
df,
df.iloc[z].set_axis(newcols, axis=1).reset_index(drop=True).where(pd.Series(z >= 0))
], axis=1)
Example on OP's data
df = pd.read_csv(io.StringIO("""Datetime,A,B,Nearest Datetime,A_nearest,B_nearest
2016-05-15 08:36:21 06:00,21,3,,,
2016-05-15 08:36:41 06:00,43,3,2016-05-15 08:36:21 06:00,21.0,3.0
2016-05-15 08:36:50 06:00,54,45,2016-05-15 08:36:21 06:00,21.0,3.0
2016-05-15 08:37:10 06:00,2,23,2016-05-15 08:36:50 06:00,54.0,45.0
2016-05-15 08:37:19 06:00,54,8,2016-05-15 08:36:50 06:00,54.0,45.0
2016-05-15 08:37:39 06:00,67,6,2016-05-15 08:37:19 06:00,54.0,8.0
"""), parse_dates=['Datetime', 'Nearest Datetime'])
>>> select_2(df[['Datetime', 'A', 'B']])
Datetime A B Datetime_nearest A_nearest B_nearest
0 2016-05-15 08:36:21 06:00 21 3 NaT NaN NaN
1 2016-05-15 08:36:41 06:00 43 3 2016-05-15 08:36:21 06:00 21.0 3.0
2 2016-05-15 08:36:50 06:00 54 45 2016-05-15 08:36:21 06:00 21.0 3.0
3 2016-05-15 08:37:10 06:00 2 23 2016-05-15 08:36:50 06:00 54.0 45.0
4 2016-05-15 08:37:19 06:00 54 8 2016-05-15 08:36:50 06:00 54.0 45.0
5 2016-05-15 08:37:39 06:00 67 6 2016-05-15 08:37:19 06:00 54.0 8.0
And on a gen(10_000) df (from the tests below):
% timeit select_2(df)
3.31 ms ± 6.79 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
>>> select(df).equals(select_2(df))
True
Original answer
IIUC, you can do it in a vectorized way, using a rolling window on column Datetime based on a time offset:
# 1. find the first row of rolling windows of 20s, left-open
# then subtract 1 (to get first row at 20s or more away from
# the current one)
z = (
df.assign(rownum=range(len(df)))
.rolling(pd.Timedelta('20s'), on='Datetime', closed='right')['rownum']
.apply(min).astype(int) - 1
)
# 2. take the rows indicated by z, make sure row index < 0
# are replaced by NaN (or NaT for Datetime), amend the column
# names and concat
cols = ['Datetime', 'A', 'B']
newcols = [f'{k}_nearest' for k in cols]
out = pd.concat([
df[cols],
df.iloc[z][cols].set_axis(newcols, axis=1).reset_index(drop=True).where(z >= 0)
], axis=1)
On your data:
>>> z
0 -1
1 0
2 0
3 2
4 2
5 4
>>> out
Datetime A B Datetime_nearest A_nearest B_nearest
0 2016-05-15 08:36:21 06:00 21 3 NaT NaN NaN
1 2016-05-15 08:36:41 06:00 43 3 2016-05-15 08:36:21 06:00 21.0 3.0
2 2016-05-15 08:36:50 06:00 54 45 2016-05-15 08:36:21 06:00 21.0 3.0
3 2016-05-15 08:37:10 06:00 2 23 2016-05-15 08:36:50 06:00 54.0 45.0
4 2016-05-15 08:37:19 06:00 54 8 2016-05-15 08:36:50 06:00 54.0 45.0
5 2016-05-15 08:37:39 06:00 67 6 2016-05-15 08:37:19 06:00 54.0 8.0
Testing
# 1. make a function with the solution above
def select(df, min_dt='20s'):
newcols = [f'{k}_nearest' for k in df.columns]
z = (
df.assign(rownum=range(len(df)))
.rolling(pd.Timedelta(min_dt), on='Datetime', closed='right')['rownum']
.apply(min).astype(int) - 1
)
return pd.concat([
df,
df.iloc[z].set_axis(newcols, axis=1).reset_index(drop=True).where(z >= 0)
], axis=1)
# 2. make a generator of test data (with just the right columns)
import numpy as np
def gen(n):
return pd.DataFrame({
'Datetime': np.random.randint(1, 30, n).cumsum() * pd.Timedelta('1s') pd.Timestamp('2020'),
'A': np.random.randint(0, 100, n),
'B': np.random.randint(0, 100, n),
})
Example
np.random.seed(0)
df = gen(10)
>>> select(df)
Datetime A B Datetime_nearest A_nearest B_nearest
0 2020-01-01 00:00:13 21 87 NaT NaN NaN
1 2020-01-01 00:00:29 36 46 NaT NaN NaN
2 2020-01-01 00:00:51 87 88 2020-01-01 00:00:29 36.0 46.0
3 2020-01-01 00:00:52 70 81 2020-01-01 00:00:29 36.0 46.0
4 2020-01-01 00:00:56 88 37 2020-01-01 00:00:29 36.0 46.0
5 2020-01-01 00:01:24 88 25 2020-01-01 00:00:56 88.0 37.0
6 2020-01-01 00:01:28 12 77 2020-01-01 00:00:56 88.0 37.0
7 2020-01-01 00:01:36 58 72 2020-01-01 00:00:56 88.0 37.0
8 2020-01-01 00:01:46 65 9 2020-01-01 00:01:24 88.0 25.0
9 2020-01-01 00:02:06 39 20 2020-01-01 00:01:46 65.0 9.0
Speed
df = gen(10_000)
%timeit select(df)
# 398 ms ± 1.04 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
%timeit select_2(df)
3.31 ms ± 6.79 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Q & A
why is this not
O[n^2]?rollingisO[n](on a sorted index), because it maintains two pointers, one for the left index of the window, one for the right. It just loops over the right index (O[n]) and increments the left index only if needed (alsoO[n]). Note that we useminas the function to apply, just for convenience. We could instead use aselect_firstto just get the first value, but that practically doesn't save much time.why use an auxiliary variable
rownumand not simply the first index of each window (aTimestamp)?That's because
rollingis rather picky at what functions it can apply. It does want a numerical value (afloat, really) and croaks if the applied function returns for example aTimestamp. In addition, it is helpful to have a simple row number, esp. in the case where several rows may have the sameDatetime.why are we subtracting 1 in the expression for
z?That is because the problem statement is: find the previous row that is at least 20 s out. The way we solve that is by taking a window that is left-open, right-closed (so exactly 20 s would be outside the window), then we subtract 1 to get the first row that is at least 20 seconds away from the current. The negative values that may appear are indicating that no such row exists, and are replaced by NaN.
why is
np.searchsorted100x faster?With
np.searchsorted, we get at the heart of the algorithm (searching the first row that is at least 20s away from the current row), also in a vectorized way in C, without having to iterate in Python (like it is done in Pandas -- seeget_window_bounds()), and without having to make sub-frames or any other overhead.
CodePudding user response:
The following solution is memory-efficient but it is not the fastest one (because it uses iteration over rows).
The fully vectorized version (that I could think of on my own) would be faster but it would use O(n^2) memory.
Example dataframe:
timestamps = [pd.Timestamp('2016-01-01 00:00:00'),
pd.Timestamp('2016-01-01 00:00:19'),
pd.Timestamp('2016-01-01 00:00:20'),
pd.Timestamp('2016-01-01 00:00:21'),
pd.Timestamp('2016-01-01 00:00:50')]
df = pd.DataFrame({'Datetime': timestamps,
'A': np.arange(10, 15),
'B': np.arange(20, 25)})
| Datetime | A | B | |
|---|---|---|---|
| 0 | 2016-01-01 00:00:00 | 10 | 20 |
| 1 | 2016-01-01 00:00:19 | 11 | 21 |
| 2 | 2016-01-01 00:00:20 | 12 | 22 |
| 3 | 2016-01-01 00:00:21 | 13 | 23 |
| 4 | 2016-01-01 00:00:50 | 14 | 24 |
Solution:
times = df['Datetime'].to_numpy() # it's convenient to have it as an `ndarray`
shifted_times = times - pd.Timedelta(20, unit='s')
usefulis a list of "useful" indices ofdf- i.e. where the appended values will NOT benan:
useful = np.nonzero(shifted_times >= times[0])[0]
# useful == [2, 3, 4]
- Truncate
shifted_timesfrom the beginning - to iterate through useful elements only:
if len(useful) == 0:
# all new columns will be `nan`s
first_i = 0 # this value will never actually be used
useful_shifted_times = np.array([], dtype=shifted_times.dtype)
else:
first_i = useful[0] # first_i == 2
useful_shifted_times = shifted_times[first_i : ]
Find the corresponding index positions of
dffor each "useful" value.(these index positions are essentially the indices of
timesthat are selected for each element ofuseful_shifted_times):
selected_indices = []
# Iterate through `useful_shifted_times` one by one:
# (`i` starts at `first_i`)
for i, shifted_time in enumerate(useful_shifted_times, first_i):
selected_index = np.nonzero(times[: i] <= shifted_time)[0][-1]
selected_indices.append(selected_index)
# selected_indices == [0, 0, 3]
- Selected rows:
df_nearest = df.iloc[selected_indices].add_suffix('_nearest')
| Datetime_nearest | A_nearest | B_nearest | |
|---|---|---|---|
| 0 | 2016-01-01 00:00:00 | 10 | 20 |
| 0 | 2016-01-01 00:00:00 | 10 | 20 |
| 3 | 2016-01-01 00:00:21 | 13 | 23 |
Replace indices of
df_nearestto match those of the corresponding rows ofdf.(basically, that is the last
len(selected_indices)indices):
df_nearest.index = df.index[len(df) - len(selected_indices) : ]
| Datetime_nearest | A_nearest | B_nearest | |
|---|---|---|---|
| 2 | 2016-01-01 00:00:00 | 10 | 20 |
| 3 | 2016-01-01 00:00:00 | 10 | 20 |
| 4 | 2016-01-01 00:00:21 | 13 | 23 |
- Append the selected rows to the original dataframe to get the final result:
new_df = df.join(df_nearest)
| Datetime | A | B | Datetime_nearest | A_nearest | B_nearest | |
|---|---|---|---|---|---|---|
| 0 | 2016-01-01 00:00:00 | 10 | 20 | NaT | nan | nan |
| 1 | 2016-01-01 00:00:19 | 11 | 21 | NaT | nan | nan |
| 2 | 2016-01-01 00:00:20 | 12 | 22 | 2016-01-01 00:00:00 | 10 | 20 |
| 3 | 2016-01-01 00:00:21 | 13 | 23 | 2016-01-01 00:00:00 | 10 | 20 |
| 4 | 2016-01-01 00:00:50 | 14 | 24 | 2016-01-01 00:00:21 | 13 | 23 |
Note: NaT stands for 'Not a Time'. It is the equivalent of nan for time values.
Note: it also works as expected even if all the last 'Datetime' - 20 sec is before the very first 'Datetime' --> all new columns will be nans.
