I'm trying to save a very long EEG line plot (>100 hours with a datapoint every 5min) as a single image with a resolution ~10x10000px so that it would be readable by human eye when zoomed in.
I tried using figsize=(10000,10) but this just gives me an error that the image is too big. Anything in the figsize acceptable range returns a compressed image that is useless for EEG reading:
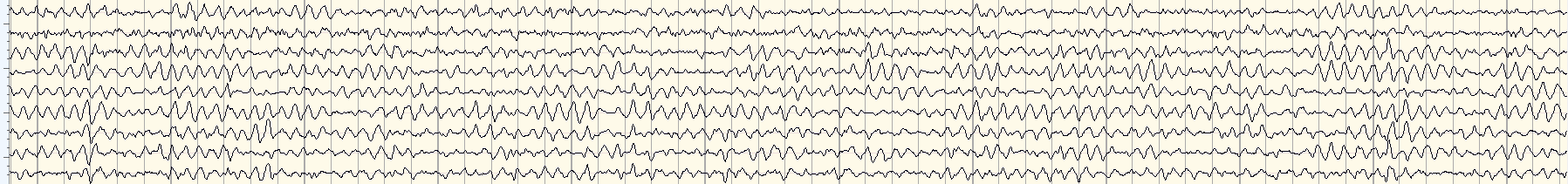
What I want instead is something like this:
UPDATE:
I've tried both increasing the dpi and using .svg as format. This is the code as requested:
def visualize_eeg(mat_path):
mat = scipy.io.loadmat(mat_path)
fs = mat['fs'][0][0]
ch_labels = [x[0].upper() for x in mat['ch_labels'][0]]
eeg_data = list(list(x) for x in mat['eeg_data'])
dict_data = dict(zip(ch_labels,eeg_data))
df = pd.DataFrame(dict_data)
df = df.T.sort_index().T
fig = plt.figure(figsize=(50, 10))
gs = gridspec.GridSpec(len(df.columns), 1,wspace=0.0, hspace=0.0)
hours = get_hours(df,fs)
# plot each column (channel) in a subplot
for i in tqdm(range(len(df.columns))):
ch = df.columns[i]
ax = plt.subplot(gs[i])
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.set_yticks([0])
ax.set_yticklabels([ch])
if i == len(df.columns)-1:
ax.tick_params(left = True, right = False , labelleft = True ,
labelbottom = True, bottom = True)
ax.set_xticks(hours.index,hours)
else:
ax.spines['bottom'].set_visible(False)
ax.tick_params(left = True, right = False , labelleft = True ,
labelbottom = False, bottom = False)
ax.plot(df[ch].apply(lambda x : x*1000), label=ch)
fig.set_dpi(1000)
fig.savefig('test.svg')
But what I get is just a scrollable crappy image (zoom or not):

CodePudding user response:
Your best bet might be to save the figure as a Scalable Vector Graphic (svg) and then just open it in Chrome or another browser (as they support svg viewing). SVGs are great because you can zoom in as much as you want without losing resolution. This figure has 10,000 points which is 8x larger than your figure (10 hours * 60 min / 5 min interval = 1200 points) and it can be viewed fine.
import matplotlib.pyplot as plt
data = [2515, 2513, 2513, 2513, 2513, 2512, 2515, 2511, 2526, 2513, 2511, 2500, 2521, 2511, 2523,
2512, 2529, 2513, 2526, 2514, 2518, 2512, 2524, 2512, 2527, 2512, 2521, 2512, 2517, 2514,
2522, 2512, 2521, 2512, 2528, 2511, 2523, 2512, 2518, 2513, 2522, 2512, 2511, 2512, 2524,
2512, 2515, 2512, 2509, 2512, 2515, 2512, 2528, 2512, 2516, 2512, 2527, 2512, 2526, 2512,
2528, 2512, 2529, 2512, 2523, 2511, 2526, 2512, 2521, 2513, 2510, 2512, 2523, 2513, 2500,
2511, 2518, 2512, 2513, 2512, 2526, 2512, 2526, 2512, 2520, 2512, 2526, 2512, 2519, 2500,
2529, 2511, 2514, 2512, 2522, 2512, 2513, 2512, 2515, 2512]*100
plt.rcParams["figure.figsize"] = (900,10)
plt.plot(data)
plt.savefig("test.svg")
CodePudding user response:
Thank you for all your answers and comments, in the end, I decided to go back to use MNE, take distinct snapshot of each 10min segment of EEG:
for seg in range(segments):
fig = raw.plot(start=seg*600, duration=600, show=False, scalings={'eeg':df.std().mean()*2},
show_scrollbars=False, show_scalebars=False)
fig.savefig('temp_subplots/%s_%i.png' % (filename,h))
plt.close()
Then I used Pillow to concatenate the images together to form a single picture of the long trace EEG:
from PIL import Image
def merge_images(filename):
images = []
all_images = [x for x in os.listdir('temp_subplots/') if filename in x]
for i in all_images:
images.append(Image.open('temp_subplots/%s' % i))
image_size = images[0].size
new_image = Image.new('RGB',(image_size[0] * len(images), image_size[1]), (250,250,250))
for i,img in enumerate(images):
new_image.paste(img,(img.size[0]*i,0))
new_image.save("long_plots/%s.png" % filename)
while not os.path.isfile("long_plots/%s.png" % filename):
sleep(1)
shutil.rmtree('temp_subplots')