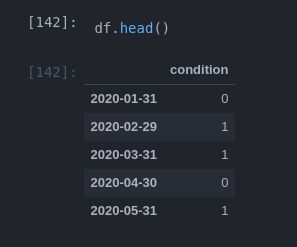
I have a data set with timeseries data. When a condition is met for a parameter I want to measure for how long that was.
I can for loop through all the positions where the condition changes but that seems to be inefficient.
What is the best way to do this vectorized?
Example:
import numpy as np
import pandas as pd
np.random.seed(0)
# generate dataset:
df = pd.DataFrame({'condition': np.random.randint(0, 2, 24)},
index = pd.date_range(start='2020', freq='M', periods=24))
df
Goal:
my goal is to create a column with the duration of sub sequential occurrences of '1' in this example:
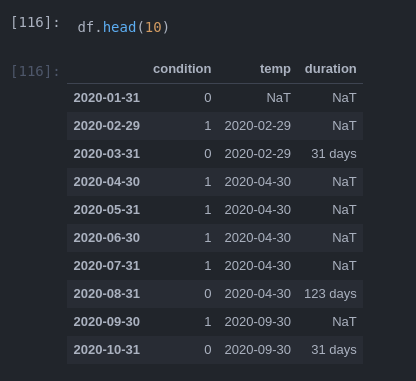
what I did so far:
# find start and end of condition:
ends = df[df.condition.diff() < 0].index
start = df[df.condition.diff() > 0].index[:ends.size]
# loop through starts and determine length
for s, e in zip(start, ends):
df.loc[e, 'duration'] = e - s
# move 1 step back so it matches with last value position
df['duration'] = df.duration.shift(-1)
in this example this is pretty fast but the loop makes it slow with bigger datasets. What would be the fastest way to do something like this?
CodePudding user response:
One way I managed to vectorize it is using 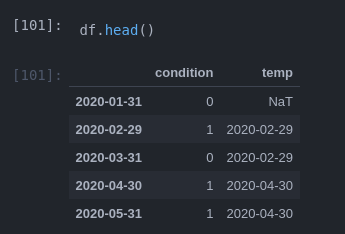
Subtract start from end:
df.loc[ends, 'duration'] = ends - df.loc[ends, 'temp']
shift 1 step back to end:
df['duration'] = df.duration.shift(-1)
This ~1000x Faster on a dataframe with 1e5 rows:
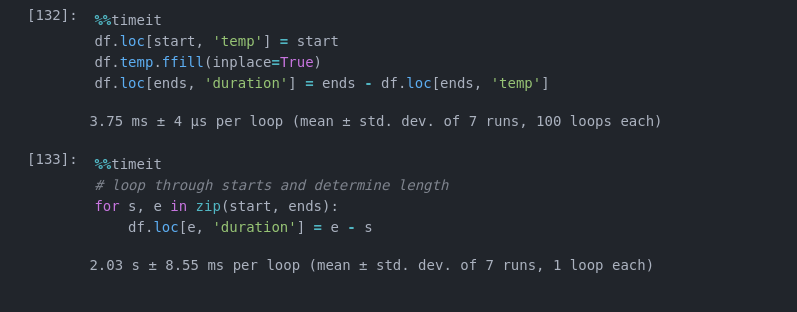
But I still wonder if this could be further improved...
CodePudding user response:
Mark the first '0' in a '0' group and mark the first '1' in a '1' group. Use .loc to select only those rows. Then do a diff on those. Use .loc to keep only the diffs for the first '0' rows. Then assign back to the original dataframe relying on the index to place the durations into the appropriate rows (almost). Then shift into proper place.
s0 = df['condition'].diff() < 0
s1 = df['condition'].diff() > 0
dfc = (
df.assign(duration=df.loc[s0|s1].index.to_series().diff().loc[s0])
.assign(duration=lambda x: x['duration'].shift(-1))
)
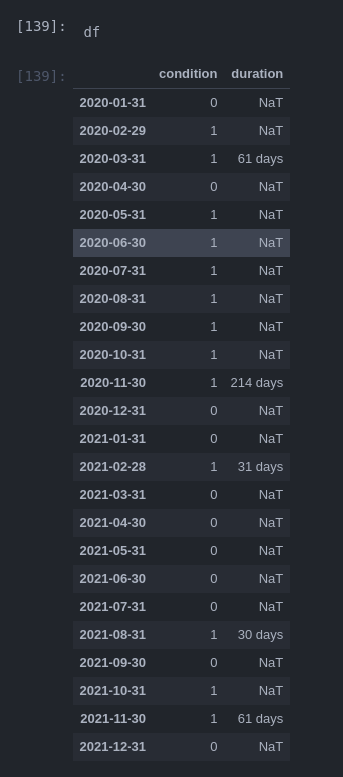
Result (see timings below this result section)
print(dfc)
condition duration
2020-01-31 0 NaT
2020-02-29 1 NaT
2020-03-31 1 61 days
2020-04-30 0 NaT
2020-05-31 1 NaT
2020-06-30 1 NaT
2020-07-31 1 NaT
2020-08-31 1 NaT
2020-09-30 1 NaT
2020-10-31 1 NaT
2020-11-30 1 214 days
2020-12-31 0 NaT
2021-01-31 0 NaT
2021-02-28 1 31 days
2021-03-31 0 NaT
2021-04-30 0 NaT
2021-05-31 0 NaT
2021-06-30 0 NaT
2021-07-31 0 NaT
2021-08-31 1 30 days
2021-09-30 0 NaT
2021-10-31 1 NaT
2021-11-30 1 61 days
2021-12-31 0 NaT
Timing DataFrame
19,135 rows with unique index
Timing - Question - n4321d
%%timeit
ends = df[df.condition.diff() < 0].index
start = df[df.condition.diff() > 0].index[:ends.size]
# loop through starts and determine length
for s, e in zip(start, ends):
df.loc[e, 'duration'] = e - s
# move 1 step back so it matches with last value position
df['duration'] = df.duration.shift(-1)
500 ms ± 23 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Timing - Answer - n4321d
%%timeit
ends = df[df.condition.diff() < 0].index
start = df[df.condition.diff() > 0].index[:ends.size]
df.loc[start, 'temp'] = start
df.temp.ffill(inplace=True)
df.loc[ends, 'duration'] = ends - df.loc[ends, 'temp']
df['duration'] = df.duration.shift(-1)
8.78 ms ± 167 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Timing - Answer - jch
%%timeit
s0 = df['condition'].diff() < 0
s1 = df['condition'].diff() > 0
dfc = (
df.assign(duration=df.loc[s0|s1].index.to_series().diff().loc[s0])
.assign(duration=lambda x: x['duration'].shift(-1))
)
5.96 ms ± 242 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)