I have a small problem trying to propoerly parse a .txt files and show its content on a QTableView. Specifically how to extract the headers of the file and show them into a QTableView.
The .txt file is composed of a first row which carries the headers, and all the other rows, which are data.
I can successfully upload the .txt file on a QTableView but for some reasons, the whole file appears under one gigantic column instead of properly parsing all the different headers and put them on the QTableView.
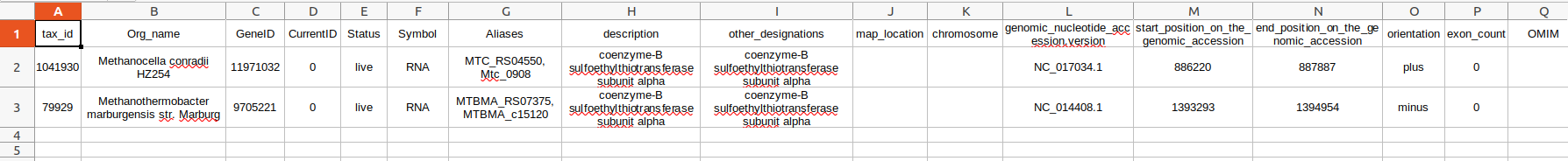
Below the example .txt file I am uploading on the QTableView - specifically the headers:
tax_id Org_name GeneID CurrentID Status Symbol Aliases description other_designations map_location chromosome genomic_nucleotide_accession.version start_position_on_the_genomic_accession end_position_on_the_genomic_accession orientation exon_count OMIM
Below the rows of some exmaple data:
1041930 Methanocella conradii HZ254 11971032 0 live mRNA MTC_RS04550, Mtc_0908 coenzyme-B sulfoethylthiotransferase subunit alpha coenzyme-B sulfoethylthiotransferase subunit alpha NC_017034.1 886220 887887 plus 0
79929 Methanothermobacter marburgensis str. Marburg 9705221 0 live mRNA MTBMA_RS07375, MTBMA_c15120 coenzyme-B sulfoethylthiotransferase subunit alpha coenzyme-B sulfoethylthiotransferase subunit alpha NC_014408.1 1393293 1394954 minus 0
523846 Methanothermus fervidus DSM 2088 9962464 0 live mRNA MFER_RS03735, Mfer_0734 coenzyme-B sulfoethylthiotransferase subunit alpha coenzyme-B sulfoethylthiotransferase subunit alpha NC_014658.1 713917 715581 plus 0
EDIT
Below the Excel file of the same .txt I posted for clarity in the visualization.
this is the expected output on the QTableView after parsing the .txt file and its headers
This is the wrong output that is currently happening which puts everything under a gigantic column.
Currenlty this upload the .txt file into the QTableView.
However it is not divided per header, but it is just a gigantic column and I don't know why despite I split the strings:
MainWindow::MainWindow(QWidget *parent)
: QMainWindow(parent)
, ui(new Ui::MainWindow)
{
ui->setupUi(this);
model = new QStandardItemModel(this);
ui->tableView->setModel(model);
ui->tableView->horizontalHeader()->setSectionResizeMode(QHeaderView::Stretch);
setWindowTitle("Viewer Example");
}
MainWindow::~MainWindow()
{
delete ui;
}
void MainWindow::on_loadTXTBtn_clicked()
{
auto filename = QFileDialog::getOpenFileName(this, "Open", QDir::rootPath(), "txt file (*.txt)");
if(filename.isEmpty()) {
return;
}
QFile file(filename);
if(!file.open(QIODevice::ReadOnly | QIODevice::Text)) {
return;
}
QTextStream xin(&file);
int ix = 0;
while (!xin.atEnd()) {
model->setRowCount(ix);
auto line = xin.readLine();
auto values = line.split(" ");
const int colCount = values.size();
model->setColumnCount(colCount);
for(int jx = 0; jx < colCount; jx) {
setValueAt(ix, jx, values.at(jx));
}
ix;
ui->pathLineEdit->setText(filename);
}
file.close();
}
void MainWindow::setValueAt(int ix, int jx, const QString &value)
{
if (!model->item(ix, jx)) {
model->setItem(ix, jx, new QStandardItem(value));
} else {
model->item(ix, jx)->setText(value);
}
}
In doing research on how to solve the problem I found this post useful and also this one. In particular the last post was very useful for understanding how to parse through the headers but I still was not able to properly understand how to extract those and show them into a QTableView.
Please any pointers on how to solve would be great!
CodePudding user response:
Assuming you have your data in cvs file with ; separator instead of .txt The code will be like this:
namespace constants
{
const QStringList HEADERS = {
"tax_id", "Org_name", "GeneID", "CurrentID", "Status",
"Symbol", "Aliases", "description", "other_designations",
"map_location", "chromosome", "genomic_nucleotide_accession.version",
"start_position_on_the_genomic_accession", "end_position_on_the_genomic_accession",
"orientation", "exon_count", "OMIM"};
}
MainWindow::MainWindow(QWidget *parent) :
QMainWindow(parent),
ui(new Ui::MainWindow)
{
ui->setupUi(this);
model = new QStandardItemModel(this);
ui->tableView->setModel(model);
model->setHorizontalHeaderLabels(constants::HEADERS);
model->setColumnCount(constants::HEADERS.length());
ui->tableView->horizontalHeader()->setSectionResizeMode(QHeaderView::ResizeMode::ResizeToContents);
setWindowTitle("Viewer Example");
OpenFile();
}
void MainWindow::OpenFile()
{
auto filename = QFileDialog::getOpenFileName(this, "Open", QDir::rootPath(), "txt file (*.csv)");
if(filename.isEmpty()) {
return;
}
QFile file(filename);
if(!file.open(QIODevice::ReadOnly | QIODevice::Text)) {
return;
}
QTextStream xin(&file);
int row = 0;
while (!xin.atEnd()) {
auto line = xin.readLine();
auto values = line.split(";");
const int colCount = model->columnCount();
for(int col = 0; col < colCount; col)
{
setValueAt(row, col, values.at(col));
}
row ;
ui->pathLineEdit->setText(filename);
}
file.close();
}
void MainWindow::setValueAt(int ix, int jx, const QString &value)
{
if (!model->item(ix, jx)) {
model->setItem(ix, jx, new QStandardItem(value));
} else {
model->item(ix, jx)->setText(value);
}
}
Your data.csv example file will be like this:
1041930;Methanocella conradii HZ254;11971032;0;live;mRNA;MTC_RS04550, Mtc_0908;coenzyme-B sulfoethylthiotransferase subunit alpha;coenzyme-B sulfoethylthiotransferase subunit alpha;;;NC_017034.1;886220;887887;plus;0;;
79929;Methanothermobacter marburgensis str. Marburg;9705221;0;live;mRNA;MTBMA_RS07375, MTBMA_c15120;coenzyme-B sulfoethylthiotransferase subunit alpha;coenzyme-B sulfoethylthiotransferase subunit alpha;;;NC_014408.1;1393293;1394954;minus;0;;
I don't think there is a clean way to achieve what you want with the way your txt file is formatted.