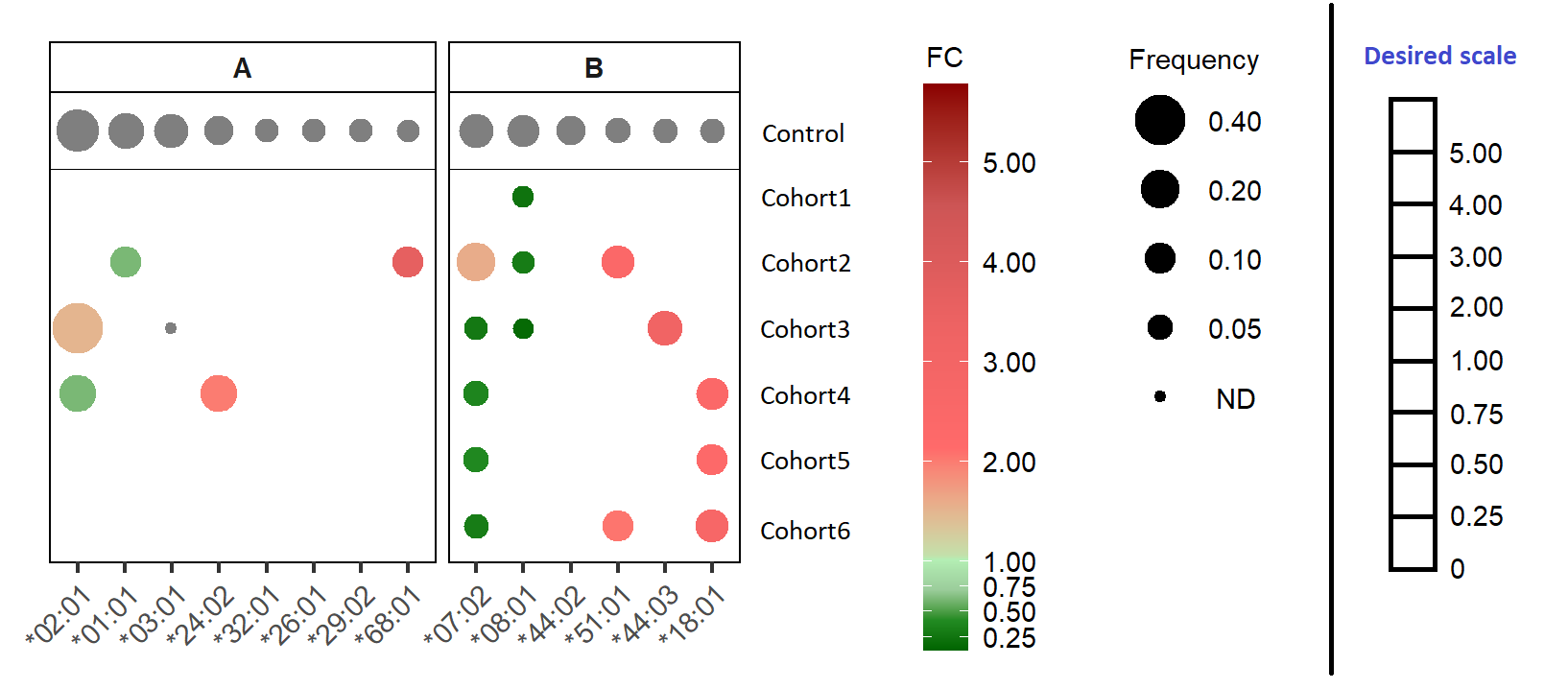
I'm plotting 2 variables (frequency and fold change) by geom_point. As in the figure below, the size corresponds to the frequency and colour to the fold change (FC <1.5 = green, >1.5 = red). To distinguish fold change below 1, I have introduced more breaks. The plot turns out as expected, but I am wondering if there is a way to make the scale at the legend to be equally spaced for the breaks corresponding to this scale c(0, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6) (see "Desired scale" on the right of the plot). Does anyone knows how to achieve this? Thanks a lot in advanced!
Below is the code for the plot.
p <- ggplot(mainG, aes(x = Allele, y = Cohort, size = Freq, color = FC))
geom_point()
scale_y_discrete(limits = rev(levels(mainG$Cohort)), position = "right")
scale_size_continuous(limits = c(0, 0.5), breaks = c(0, 0.05, 0.10, 0.20, 0.40))
# Underrepresented: FC < 1.5 ; overrepresented: FC > 1.5
scale_colour_gradientn(
colours = c('darkgreen', 'forestgreen', 'darkseagreen3', 'darkseagreen2',
'indianred1', 'indianred2', 'indianred3', 'darkred'),
values = c(0, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6)/6,
breaks = c(0, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6))
xlab("") ylab("")
theme(axis.text = element_text(size = 7),
axis.text.x = element_text(angle = 45, vjust = 1, hjust=1),
axis.ticks.y = element_blank(),
panel.border = element_rect(fill = NA),
panel.background = element_blank(),
axis.line = element_line(),
legend.title = element_text(size = 7),
legend.text = element_text(size = 7),
legend.key = element_blank(),
legend.position = "right",
panel.spacing = unit(0.2, "lines"),
strip.background =element_rect(colour = "black", fill = NA),
strip.text = element_text(size = 7, face = "bold", margin = margin(0.1,5,0.1,5, "cm"))
)
facet_grid(.~ Gene, scales = "free", space = "free")
p guides(colour = guide_colourbar(barwidth = unit(0.5, "cm"), barheight = unit(5, "cm"),
direction = "vertical"),
size = guide_legend(title = "Frequency", reverse = T))
Edit: here's the sample data of mainG for reproducibility (sorry the section is becoming lengthy...)
Cohort Gene Allele Freq FC
Cohort1 B *08:01 0.027 0.24
Cohort2 A *01:01 0.103 0.63
Cohort2 A *68:01 0.103 3.63
Cohort2 B *07:02 0.207 1.59
Cohort2 B *08:01 0.034 0.31
Cohort2 B *51:01 0.121 2.44
Cohort3 A *02:01 0.407 1.51
Cohort3 A *03:01 0 NA
Cohort3 B *07:02 0.037 0.28
Cohort3 B *08:01 0.019 0.17
Cohort3 B *44:03 0.148 3.15
Cohort4 A *02:01 0.17 0.63
Cohort4 A *24:02 0.17 2.01
Cohort4 B *07:02 0.05 0.38
Cohort4 B *18:01 0.11 2.41
Cohort5 B *07:02 0.053 0.4
Cohort5 B *18:01 0.105 2.31
Cohort6 B *07:02 0.041 0.31
Cohort6 B *18:01 0.122 2.69
Cohort6 B *51:01 0.102 2.06
Control A *01:01 0.163 NA
Control A *02:01 0.269 NA
Control A *03:01 0.14 NA
Control A *24:02 0.085 NA
Control A *26:01 0.035 NA
Control A *29:02 0.035 NA
Control A *32:01 0.037 NA
Control A *68:01 0.029 NA
Control B *07:02 0.13 NA
Control B *08:01 0.11 NA
Control B *18:01 0.046 NA
Control B *44:02 0.087 NA
Control B *44:03 0.047 NA
Control B *51:01 0.05 NA
CodePudding user response:
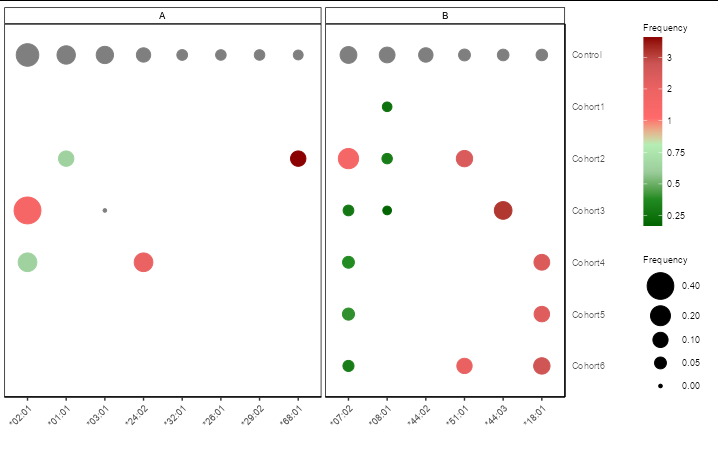
You can do this the same way as you would use a secondary axis: transform the data and apply the inverse transformation in the scale:
ggplot(mainG, aes(x = Allele, y = Cohort, size = Freq,
color = ifelse(FC < 1, FC * 4, FC 3)))
geom_point()
scale_y_discrete(limits = rev(levels(mainG$Cohort)), position = "right")
scale_size_continuous(limits = c(0, 0.5), range = c(1, 10),
breaks = c(0, 0.05, 0.10, 0.20, 0.40))
scale_colour_gradientn(name = "Frequency",
colours = c('darkgreen', 'forestgreen', 'darkseagreen3', 'darkseagreen2',
'indianred1', 'indianred2', 'indianred3', 'darkred'),
values = 0:10 / 10, breaks = 0:10, labels = ~ifelse(.x < 4, .x/4, .x-3))
xlab("") ylab("")
theme(axis.text = element_text(size = 7),
axis.text.x = element_text(angle = 45, vjust = 1, hjust=1),
axis.ticks.y = element_blank(),
panel.border = element_rect(fill = NA),
panel.background = element_blank(),
axis.line = element_line(),
legend.title = element_text(size = 7),
legend.text = element_text(size = 7),
legend.key = element_blank(),
legend.position = "right",
panel.spacing = unit(0.2, "lines"),
strip.background =element_rect(colour = "black", fill = NA),
strip.text = element_text(size = 7, face = "bold",
margin = margin(0.1,5,0.1,5, "cm")))
facet_grid(.~ Gene, scales = "free", space = "free")
guides(colour = guide_colourbar(barwidth = unit(0.5, "cm"),
barheight = unit(5, "cm"),
direction = "vertical"),
size = guide_legend(title = "Frequency", reverse = T))
CodePudding user response:
Using logs one gets close to what the question asks without confusing those reading the plot by using a non-uniform scale. It is fairly easy to modify this example to use other log bases, other transformations or other colours. Alternatively, the transformation can be applied in the scale rather than in aes(). (I did not include the call to theme() as it is not relevant to the question.)
library(ggplot2)
ggplot(mainG, aes(x = Allele, y = Cohort, size = Freq, color = log2(FC)))
geom_point()
scale_y_discrete(limits = rev(levels(mainG$Cohort)), position = "right")
scale_size_continuous(limits = c(0, 0.5),
breaks = rev(c(0, 0.05, 0.10, 0.20, 0.40)))
scale_colour_gradient2(name = "FC",
high = "darkred", mid = "lightyellow3", low = "darkgreen",
labels = function(x) {2^x},
breaks = log2(c(c(1/8, 1/4, 1/2, 1, 2, 4, 8))))
expand_limits(colour = log2(c(1/5, 5)))
facet_grid(.~ Gene, scales = "free", space = "free")
theme_bw()
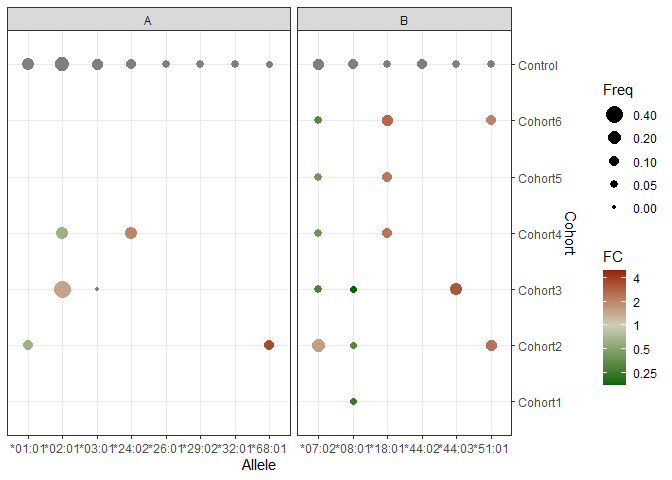
Created on 2022-09-03 with reprex v2.0.2
I think the best approach is to use one of the new binned scales for continuous data from 'ggploy2'.
library(ggplot2)
ggplot(mainG, aes(x = Allele, y = Cohort, size = Freq, color = log2(FC)))
geom_point()
scale_y_discrete(limits = rev(levels(mainG$Cohort)), position = "right")
scale_size_continuous(limits = c(0, 0.5),
breaks = rev(c(0, 0.05, 0.10, 0.20, 0.40)))
scale_colour_steps2(name = "FC",
high = "darkred", mid = "lightyellow3", low = "darkgreen",
labels = function(x) {2^x},
breaks = log2(c(c(1/8, 1/4, 1/2, 1, 2, 4, 8))))
expand_limits(colour = log2(c(1/5, 5)))
facet_grid(.~ Gene, scales = "free", space = "free")
theme_bw()
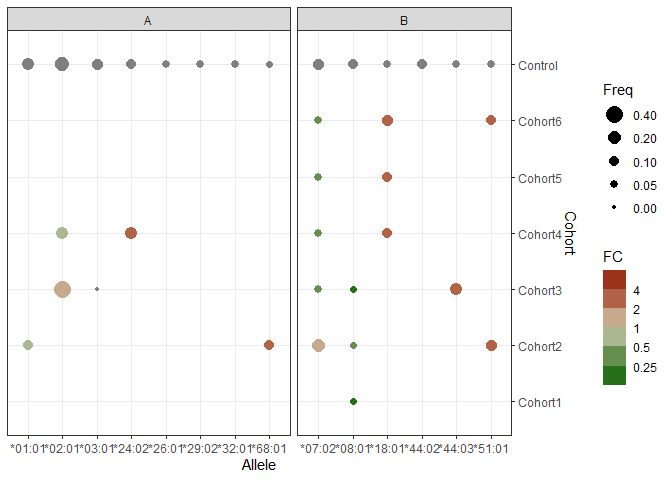
Created on 2022-09-03 with reprex v2.0.2