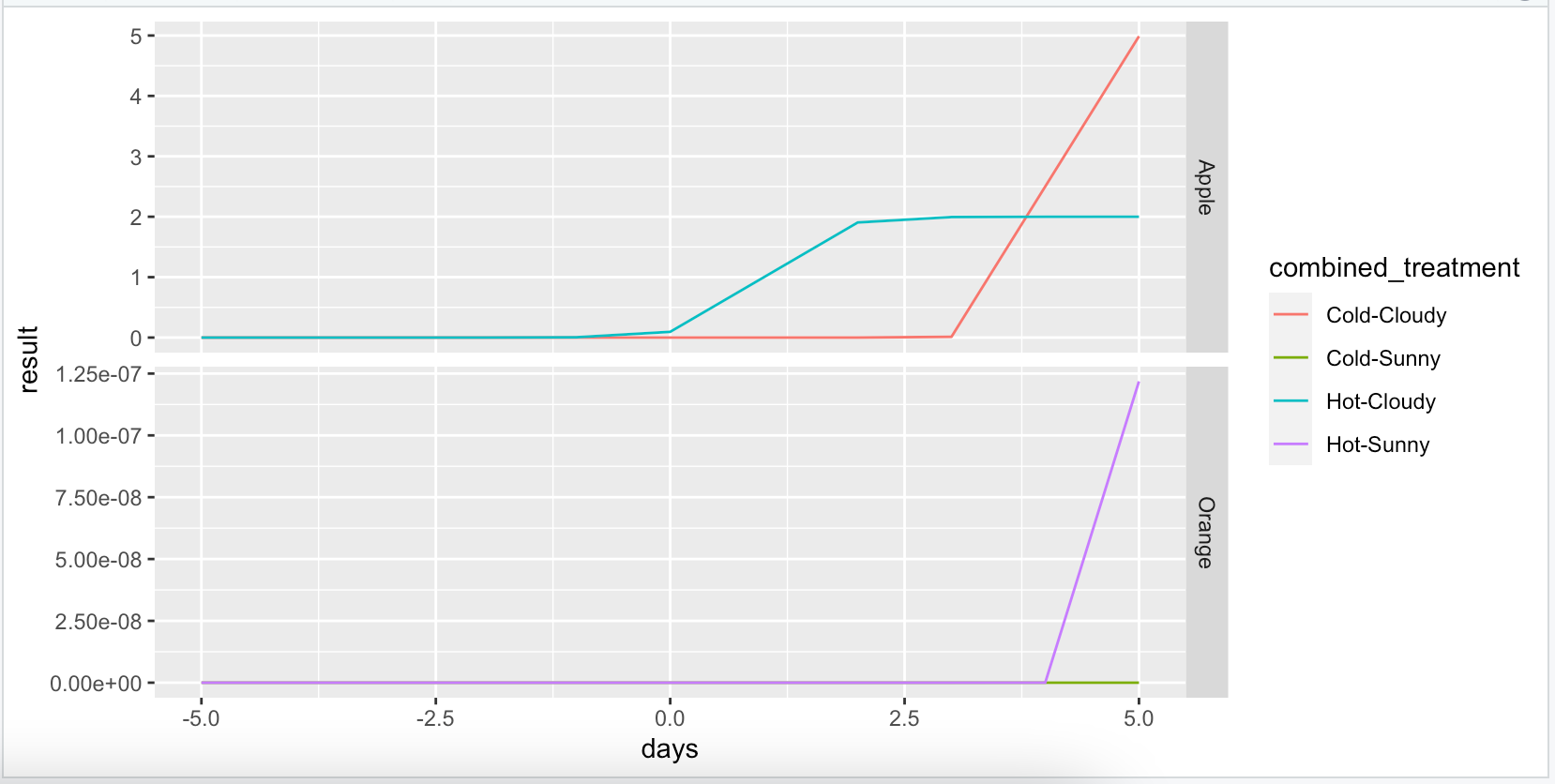
I have a dataset (test_df) that looks like:
| Species | TreatmentA | TreatmentB | X0 | L | K |
|---|---|---|---|---|---|
| Apple | Hot | Cloudy | 1 | 2 | 3 |
| Apple | Cold | Cloudy | 4 | 5 | 6 |
| Orange | Hot | Sunny | 7 | 8 | 9 |
| Orange | Cold | Sunny | 10 | 11 | 12 |
I would like to display the effect of the treatments by using the X0, L, and K values as coefficients in a standard logistic function and plotting the same species across various treatments on the same plot. I would like a grid of plots with the logistic curves for each species on it's own plots, with each treatment then being grouped by color within every plot. In the above example, Plot1.Grid1 would have 2 logistic curves corresponding to Apple Hot and Apple Cold, and plot1.Grid2 would have 2 logistic curves corresponding to Orange Hot and Orange Cold.
The below code will create a single logistic function curve which can then be layered, but manually adding the layers for multiple treatments is tedious.
testx0 <- 1
testL <- 2
testk <- 3
days <- seq(from = -5, to = 5, by = 1)
functionmultitest <- function(x,testL,testK,testX0) {
(testL)/(1 exp((-1)*(testK) *(x - testX0)))
}
ggplot() aes(x = days, y = functionmultitest(days,testL,testk,testx0)) geom_line()