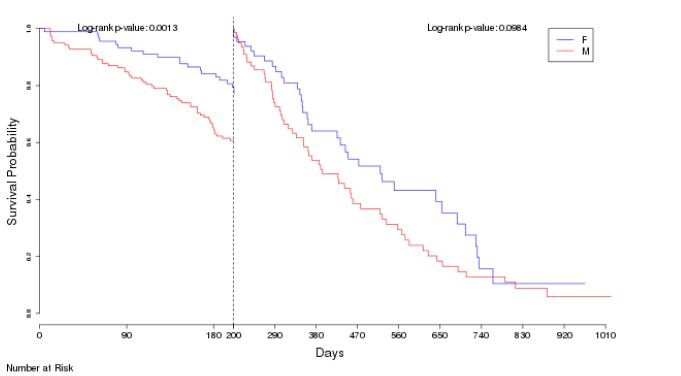
I have 5 different survfit() plots of different models that I am trying to combine into one plot in the style of a landmark analysis plot, as seen below.
At the moment they are just plot(survfit(model, newdata = )), how could I combine them so that I have the line of days 0-100 of survfit 1, 100-200 of survfit 2 etc.
CodePudding user response:
Let's create a model using the built-in lung data from the survival package:
library(survival)
library(tidyverse)
mod1 <- survfit(Surv(time, status) ~ sex, data = lung)
This model actually contains all we need to make the plot. We can convert it to a data frame as follows:
df <- as.data.frame(unclass(mod1)[c(2:7, 15:16)])
df$sex <- rep(c("Male", "Female"), times = mod1$strata)
head(df)
#> time n.risk n.event n.censor surv std.err lower upper sex
#> 1 11 138 3 0 0.9782609 0.01268978 0.9542301 1.0000000 Male
#> 2 12 135 1 0 0.9710145 0.01470747 0.9434235 0.9994124 Male
#> 3 13 134 2 0 0.9565217 0.01814885 0.9230952 0.9911586 Male
#> 4 15 132 1 0 0.9492754 0.01967768 0.9133612 0.9866017 Male
#> 5 26 131 1 0 0.9420290 0.02111708 0.9038355 0.9818365 Male
#> 6 30 130 1 0 0.9347826 0.02248469 0.8944820 0.9768989 Male
With a bit of data manipulation, we can define 100-day periods and renormalize the curves at the start of each period. Then we can plot using geom_step
df %>%
filter(time < 300) %>%
group_by(sex) %>%
mutate(period = factor(100 * floor(time / 100))) %>%
group_by(sex, period) %>%
mutate(surv = surv / first(surv)) %>%
ggplot(aes(time, surv, color = sex, group = interaction(period, sex)))
geom_step(size = 1)
geom_vline(xintercept = c(0, 100, 200, 300), linetype = 2)
scale_color_manual(values = c("deepskyblue4", "orange"))
theme_minimal(base_size = 16)
theme(legend.position = "top")
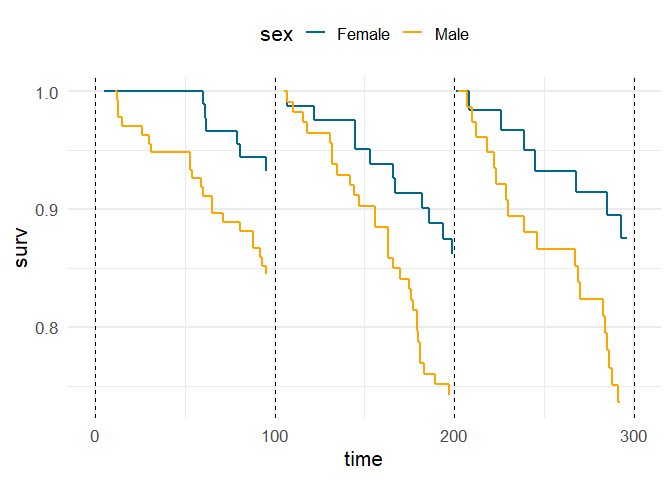
Created on 2022-09-03 with reprex v2.0.2