I have count data of invertebrates along a transect line. The data includes 3 columns- one column for the date the data was collected on, one for the transect identification number, and one for the species observed.
structure(list(Date = c("8/22/2022", "8/22/2022", "8/23/2022",
"8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022",
"8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022",
"8/23/2022", "8/23/2022", "8/18/2022", "8/18/2022", "8/18/2022",
"8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022",
"8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022",
"8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022",
"8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022", "8/23/2022",
"8/23/2022", "8/23/2022", "8/22/2022", "8/22/2022", "8/22/2022",
"8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022",
"8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022",
"8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022",
"8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022", "8/22/2022",
"8/22/2022", "8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022",
"8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022", "8/18/2022",
"8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022",
"8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022",
"8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022",
"8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022", "8/16/2022",
"8/16/2022", "8/16/2022", "8/16/2022"), Transect = c(8L, 8L,
4L, 4L, 5L, 5L, 5L, 6L, 6L, 7L, 8L, 8L, 9L, 9L, 9L, 4L, 4L, 4L,
5L, 5L, 6L, 6L, 7L, 8L, 8L, 9L, 9L, 9L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 3L, 3L,
3L, 3L, 4L, 4L, 5L, 5L, 6L, 6L, 7L, 7L, 8L, 9L, 9L, 9L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 3L, 3L, 1L, 1L, 1L, 2L, 2L, 2L, 3L, 3L, 3L,
4L, 4L, 4L, 5L, 6L, 7L, 7L, 7L, 8L, 8L, 8L, 9L, 9L, 9L), Species = c("RCRAB",
"DOL", "DOL", "STAR", "DOL", "RCRAB", "STAR", "DOL", "LOB", "DOL",
"DOL", "RCRAB", "DOL", "LOB", "STAR", "DOL", "LOB", "STAR", "DOL",
"RCRAB", "DOL", "RCRAB", "DOL", "DOL", "RCRAB", "DOL", "STAR",
"RCRAB", "DOL", "STAR", "RCRAB", "URCH", "DOL", "RCRAB", "URCH",
"STAR", "DOL", "LOB", "STAR", "URCH", "DOL", "RCRAB", "STAR",
"URCH", "STAR", "DOL", "URCH", "RCRAB", "DOL", "STAR", "URCH",
"RCRAB", "DOL", "STAR", "DOL", "LOB", "DOL", "RCRAB", "DOL",
"RCRAB", "DOL", "DOL", "STAR", "URCH", "DOL", "STAR", "RCRAB",
"LOB", "DOL", "STAR", "RCRAB", "DOL", "LOB", "DOL", "STAR", "LOB",
"DOL", "STAR", "URCH", "DOL", "STAR", "RCRAB", "DOL", "LOB",
"STAR", "DOL", "DOL", "DOL", "RCRAB", "STAR", "STAR", "DOL",
"RCRAB", "DOL", "STAR", "RCRAB")), class = "data.frame", row.names = c(NA,
-96L))
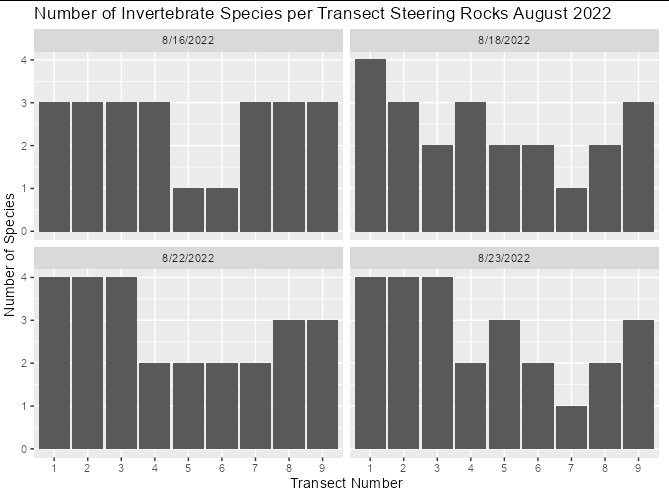
I want to create a multiplot where each date has a separate plot, with the X value being the transect number and the Y value being the number of species found on that transect. So far, I have this:
library(ggplot2)
invertplot <- ggplot(data=invert, aes(Transect, Species))
geom_bar(stat='identity')
labs(title="Number of Invertebrate Species per Transect Steering Rocks August 2022",
y="Number of Species",
x="Transect Number")
facet_wrap(~Date)
invertplot
Which gives me a plot where each individual species is listed on the Y axis, and the X axis is the number of that species in the entire data set.
How do I get ggplot to group the values by transect number, and not species? Thanks in advance!
CodePudding user response:
If you want a bar plot that simply counts observations, don't use stat = "identity". The default behaviour of geom_bar is to use stat_count. So I think you just need:
library(tidyverse)
ggplot(data = invert, aes(factor(Transect)))
geom_bar()
labs(title = paste0("Number of Invertebrate Species per Transect ",
"Steering Rocks August 2022"),
y = "Number of Species",
x = "Transect Number")
facet_wrap( ~ Date)
In case there are multiple recordings of the same species on the same transect on the same date that you only want to count once, you would be safer to do:
ggplot(data = invert %>% count(Transect, Species, Date), aes(factor(Transect)))
geom_bar()
labs(title = paste0("Number of Invertebrate Species per Transect ",
"Steering Rocks August 2022"),
y = "Number of Species",
x = "Transect Number")
facet_wrap( ~ Date)
But this gives the same output for your current example data.