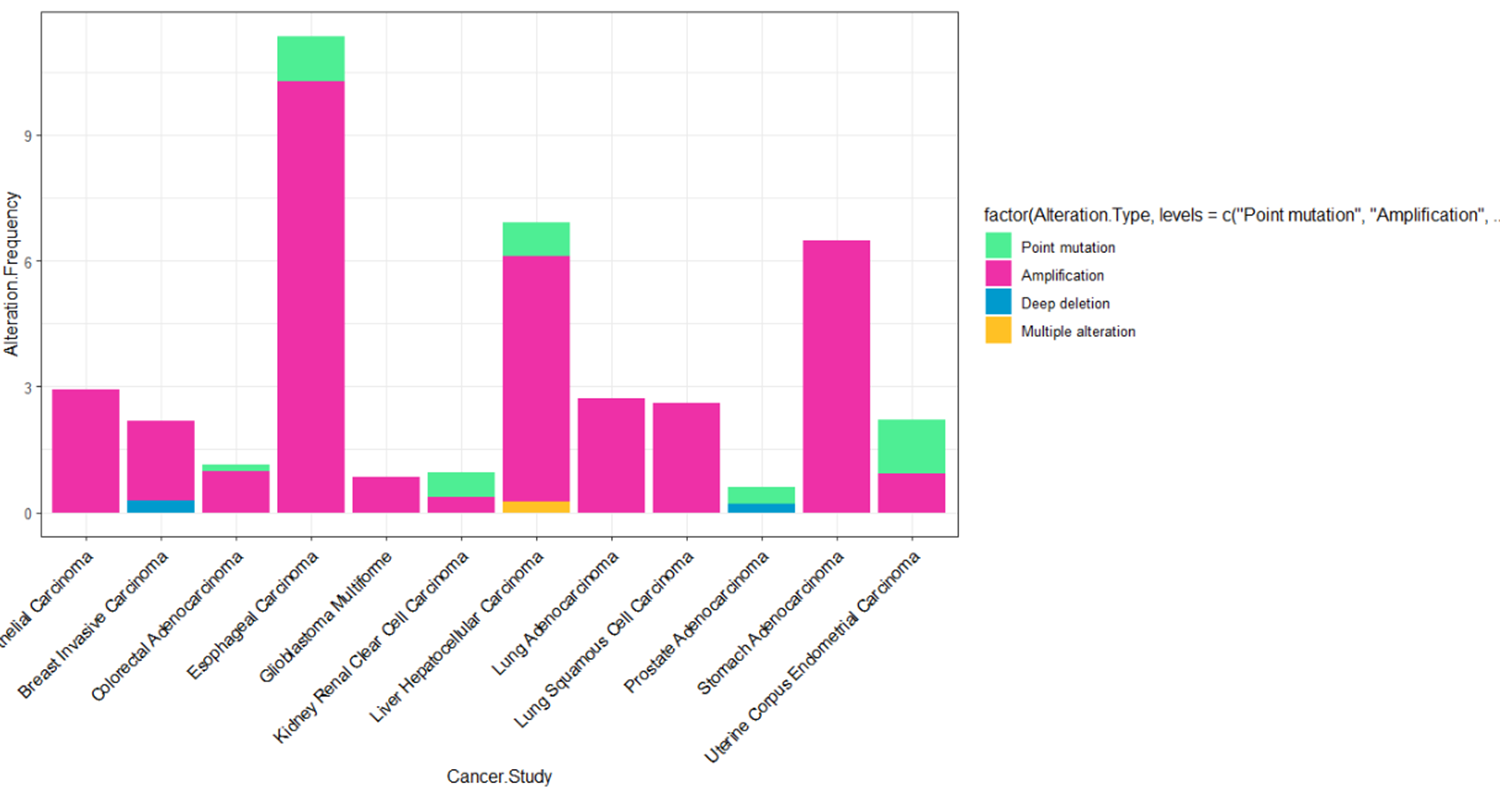
I'm trying to reorder the stacked bar plot in decreasing order, I mean the bar of Esophageal placed first, but my script doesn't work.
Cancer.Study Alteration.Frequency Alteration.Type Alteration.Count
1 Esophageal Carcinoma 10.2702703 Amplification 19
2 Esophageal Carcinoma 1.0810811 Point mutation 2
3 Liver Hepatocellular Carcinoma 0.2652520 Multiple alteration 1
4 Liver Hepatocellular Carcinoma 5.8355438 Amplification 22
5 Liver Hepatocellular Carcinoma 0.7957560 Point mutation 3
6 Stomach Adenocarcinoma 6.4853556 Amplification 31
7 Bladder Urothelial Carcinoma 2.9197080 Amplification 12
8 Lung Adenocarcinoma 2.7131783 Amplification 14
9 Lung Squamous Cell Carcinoma 2.5948104 Amplification 13
10 Uterine Corpus Endometrial Carcinoma 0.9174312 Amplification 5
And here is my script
ggplot(df,
aes(fill=factor(Alteration.Type,
levels = c('Point mutation','Amplification','Deep deletion', 'Multiple alteration')),
x=reorder(Cancer.Study, -Alteration.Frequency)), y=Alteration.Frequency))
geom_bar(position="stack", stat="identity") theme_bw()
Thanks for any help
CodePudding user response:
You could use reorder for your fill aesthetic like this:
df <- read.table(text = " Cancer.Study Alteration.Frequency Alteration.Type Alteration.Count
1 Esophageal.Carcinoma 10.2702703 Amplification 19
2 Esophageal.Carcinoma 1.0810811 Point.mutation 2
3 Liver.Hepatocellular.Carcinoma 0.2652520 Multiple.alteration 1
4 Liver.Hepatocellular.Carcinoma 5.8355438 Amplification 22
5 Liver.Hepatocellular.Carcinoma 0.7957560 Point.mutation 3
6 Stomach.Adenocarcinoma 6.4853556 Amplification 31
7 Bladder.Urothelial.Carcinoma 2.9197080 Amplification 12
8 Lung.Adenocarcinoma 2.7131783 Amplification 14
9 Lung.Squamous.Cell.Carcinoma 2.5948104 Amplification 13
10 Uterine.Corpus.Endometrial.Carcinoma 0.9174312 Amplification 5", header = TRUE)
library(ggplot2)
ggplot(df, aes(fill=reorder(Alteration.Type, -Alteration.Frequency),
x=Cancer.Study, y=Alteration.Frequency))
geom_bar(position="stack", stat="identity")
labs(fill = "Alteration.Type")
theme_bw()
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
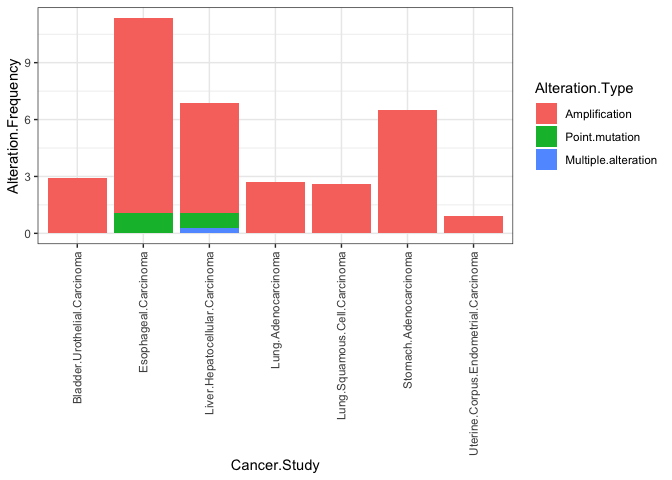
Created on 2022-09-10 with reprex v2.0.2
Please note: I added some dots to your whitespaces in your data.
CodePudding user response:
By default, reorder uses the respective mean. You however want to order by the absolute count. That's why you have to specify FUN = sum in the reoder() function.
See below with the df table of Quinten
library(tidyverse)
ggplot(df, aes(x = reorder(Cancer.Study, -Alteration.Frequency, FUN = sum),
y = Alteration.Frequency,
fill = Alteration.Type))
geom_bar(position="stack", stat="identity")
labs(fill = "Alteration.Type")
ylab("Alteration Frequency")
xlab("Cancer Study")
theme_bw()
theme(axis.text.x = element_text(angle = 45, hjust=1))