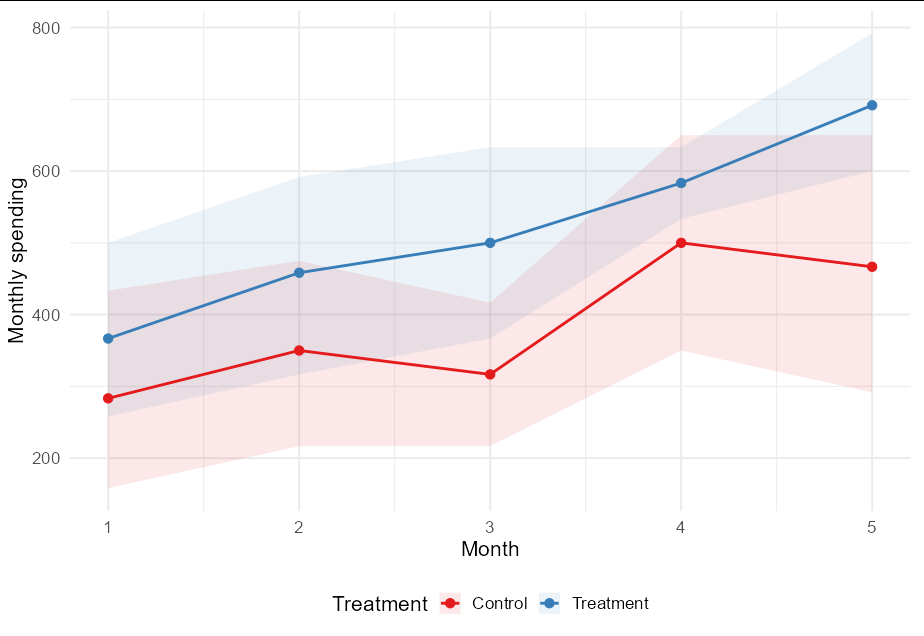
Hi I matched a sample of individuals from the treatment group and the control group by using the genetic matching approach from the MatchIt package. The outcome variables is the average spending of each month. Now I'm trying to generate a line graph to demonstrate patterns of average monthly spending of the treatment and control groups. I would like to add shaded areas to show the bootstrap confidence interval for these lines. The main issue that I encountered include:
1. Bootstrap in pairs
Bootstrap resampling for matched sample should be conducted based on each matched pair, not each individual. Thankfully I've got the solution from 
However, I don't know how to add shaded areas around these two lines to indicate the bootstrap confidence intervals from the code that I provided above. Alternatively, I could run the code for each time point again and again, and then document that in the dataset manually to plot the intervals on the graphs. However, I wonder if there's a way to conduct it more efficiently. I would be really grateful for your help.
CodePudding user response:
This seems like a very complex way to achieve your end goal. The ggplot2 function mean-cl_boot takes an input vector and returns y, ymin and ymax values calculated by a fast bootstrap method. It works directly inside summarize too, so we can replace your whole code with:
df %>%
pivot_longer(starts_with("Month"), names_to = "Month") %>%
group_by(Month, Treatment) %>%
summarize(mean_cl_boot(value)) %>%
mutate(Month = as.numeric(sub("Month_", "", Month)),
Treatment = c("Control", "Treatment")[Treatment 1]) %>%
ggplot(aes(Month, y, color = Treatment))
geom_ribbon(aes(fill = Treatment, ymin = ymin, ymax = ymax),
alpha = 0.1, color = NA)
geom_line(size = 1)
geom_point(size = 3)
scale_fill_brewer(palette = "Set1")
scale_color_brewer(palette = "Set1")
labs(y = "Monthly spending")
theme_minimal(base_size = 16)
theme(legend.position = "bottom")