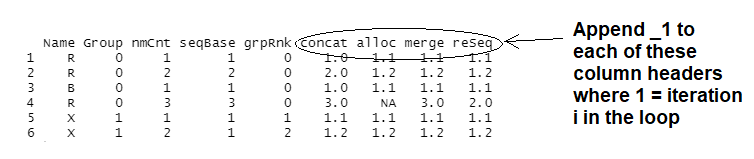
This is my first-time trying a for-loop in R. I am trying to append to the looped-over data frame column header names, the loop iteration count as shown below in the image. Columns concat, alloc, merge, reSeq, are generated in the loop and I'd like to change their names to concat_1, alloc_1, merge_1, reSeq_1 for the first loop, etc. Any recommendations for how to do this?
I'm starting very easy, looping only 1 time so I can gently step into this. In my next step I'll expand the loop so that columns are added to the right and similarly sequentially renamed. I did attempt to append to column names in the first mutate under the for-loop in the below code for each reference to concat but it doesn't work: mutate(paste0("concat_",i) = as.numeric... So I imagine the solution will be something like running colnames(...) at the bottom of the loop, etc.
Here's the code with example DF:
library(dplyr)
myDF1 <- data.frame(
Name = c("R","R","B","R","X","X"),
Group = c(0,0,0,0,1,1))
nCode <- myDF1 %>%
group_by(Name) %>%
mutate(nmCnt = row_number()) %>%
ungroup() %>%
mutate(seqBase = ifelse(Group == 0 | Group != lag(Group), nmCnt,0)) %>%
mutate(seqBase = na_if(seqBase, 0)) %>%
group_by(Name) %>%
fill(seqBase) %>%
mutate(seqBase = match(seqBase, unique(seqBase))) %>%
ungroup %>%
mutate(grpRnk = ifelse(Group > 0, sapply(1:n(), function(x) sum(Name[1:x]==Name[x] & Group[1:x] == Group[x])),0))
loopCntr <- nrow(unique(myDF1[myDF1$Group!=0,]))
for(i in 1:1) {nCode <- nCode %>%
mutate(concat = as.numeric(paste0(seqBase,".",grpRnk)))
index <- filter(nCode, Group !=0) %>%
select(concat) %>%
distinct() %>%
mutate(truncInd = trunc(concat)) %>%
group_by(truncInd) %>%
mutate(cumGrp = cur_group_id()) %>%
ungroup() %>%
select(-truncInd)
index <- if(ifelse(loopCntr > 0, min(index$concat), Inf) >= 2){
rbind(data.frame(concat=c(1),cumGrp=c(1)),index)}else{index}
nCode <- nCode %>%
mutate(alloc = index$concat[index$cumGrp==1][nmCnt]) %>%
mutate(merge = ifelse(is.na(alloc),seqBase,alloc)) %>%
group_by(Name) %>%
mutate(reSeq = match(trunc(merge), unique(trunc(merge)))) %>%
mutate(reSeq = (reSeq round(merge%%1 * 10,0)/10)) %>%
ungroup()
} # end for-loop
print.data.frame(nCode)
CodePudding user response:
Perhaps, assign with names<- or setNames or use rename_with at the end of the loop
library(dplyr)
library(stringr)
for(i in 1:1) {nCode <- nCode %>%
mutate(concat = as.numeric(paste0(seqBase,".",grpRnk)))
index <- filter(nCode, Group !=0) %>%
select(concat) %>%
distinct() %>%
mutate(truncInd = trunc(concat)) %>%
group_by(truncInd) %>%
mutate(cumGrp = cur_group_id()) %>%
ungroup() %>%
select(-truncInd)
index <- if(ifelse(loopCntr > 0, min(index$concat), Inf) >= 2){
rbind(data.frame(concat=c(1),cumGrp=c(1)),index)}else{index}
nCode <- nCode %>%
mutate(alloc = index$concat[index$cumGrp==1][nmCnt]) %>%
mutate(merge = ifelse(is.na(alloc),seqBase,alloc)) %>%
group_by(Name) %>%
mutate(reSeq = match(trunc(merge), unique(trunc(merge)))) %>%
mutate(reSeq = (reSeq round(merge%%1 * 10,0)/10)) %>%
ungroup()
nCode <- nCode %>%
rename_with(~ str_c(.x, "_", i), c("concat", "alloc", "merge", "reSeq"))
} # end for-loop
-output
> nCode
# A tibble: 6 × 9
Name Group nmCnt seqBase grpRnk concat_1 alloc_1 merge_1 reSeq_1
<chr> <dbl> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 R 0 1 1 0 1 1.1 1.1 1.1
2 R 0 2 2 0 2 1.2 1.2 1.2
3 B 0 1 1 0 1 1.1 1.1 1.1
4 R 0 3 3 0 3 NA 3 2
5 X 1 1 1 1 1.1 1.1 1.1 1.1
6 X 1 2 1 2 1.2 1.2 1.2 1.2
Or another option is using := while assignment
...
mutate(!! paste0("alloc_", i) := index$concat[index$cumGrp==1][nmCnt])%>%
...