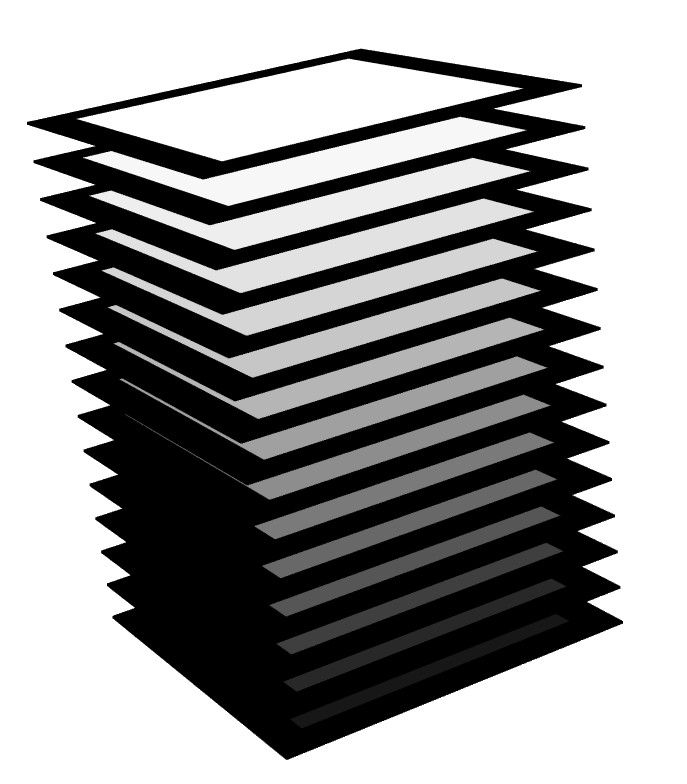
I'm trying to visualise 3D array slices (i.e. image stacks). Surprisingly, I couldn't find any package with an in-built function that easily does the job.
I provided an example code with my own solution for some dummy data but I was wondering if there is a much easier and computationally less expensive workaround? Some large computed tomography (CT) scan data takes quite a toll on my machine.
Example code:
library(leaflet) #color palette
library(rgl) #3d visualisation
#example data
slides = array(data = 0,
dim = c(200,200,15))
#add some intensity values
for(i in 1:dim(slides)[3]) slides[20:180,20:180,i] = i*10
#voxel dimensions for x/y/z
dims = c(1,1,20) #adjust z dimension for visualisation
#all possible x/y/z combinations
xyz = expand.grid(1:dim(slides)[1],1:dim(slides)[2],1:dim(slides)[3])
#find the intensity value for each data point
value = apply(xyz, 1, function(x) slides[x[1],x[2],x[3]])
#invert values
value = max(value) - value
#apply the voxel dimensions to x/y/z
xyz = t(t(xyz) * dims)
#convert intensity values to colors
colb = colorNumeric(palette = "Greys",
domain = c(0,max(value)))
col = colb(value)
#plot image stack
plot3d(xyz, col = col, aspect = FALSE, axes = FALSE, xlab = "", ylab = "", zlab = "")
Result:
CodePudding user response:
It would be much faster in rgl to use textures instead of plotting points. For example:
library(rgl) #3d visualisation
#example data
slides = array(data = 0,
dim = c(200,200,15))
#add some intensity values
n <- dim(slides)[3]
for (i in 1:n) slides[20:180,20:180,i] = i*10
# Now plot each slide to a separate file to use as a texture
filenames <- character(n)
for (i in 1:n) {
filenames[i] <- tempfile(fileext = ".png")
png(filenames[i], width=200, height=200)
raster <- as.raster(slides[,,i], max = max(slides))
par(mar = c(0,0,0,0))
plot(raster)
dev.off()
}
# Now plot one 3D quad per slide
open3d()
#> glX
#> 1
texturequad <- cbind(c(0, 1, 1, 0), c(0, 0, 1, 1))
quad <- cbind(texturequad*200, 0)
for (i in 1:n) {
quad[,3] <- i
quads3d(quad, texcoords = texturequad, texture = filenames[i],
col = "white", lit = FALSE)
}
# Set the shape of the display as desired
aspect3d(1,1,2)
lowlevel() # Just to get reprex to show it
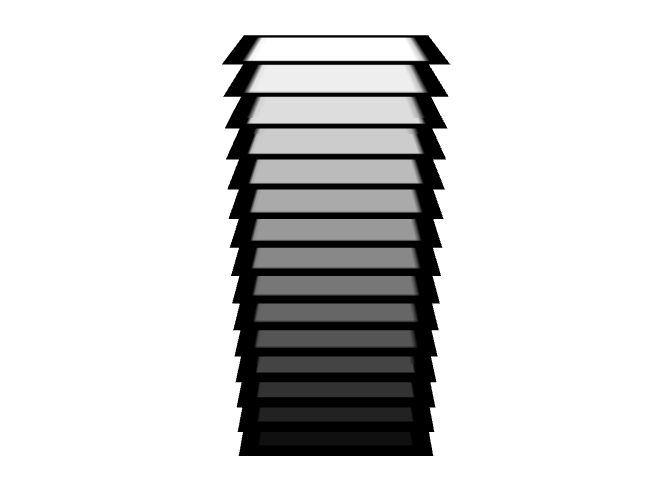
Created on 2022-09-27 with reprex v2.0.2
I didn't use your palette; if you want that, you should set the entries of slides to color values instead of numeric values.