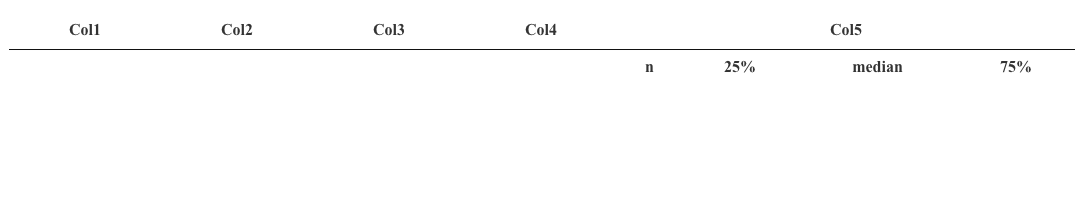
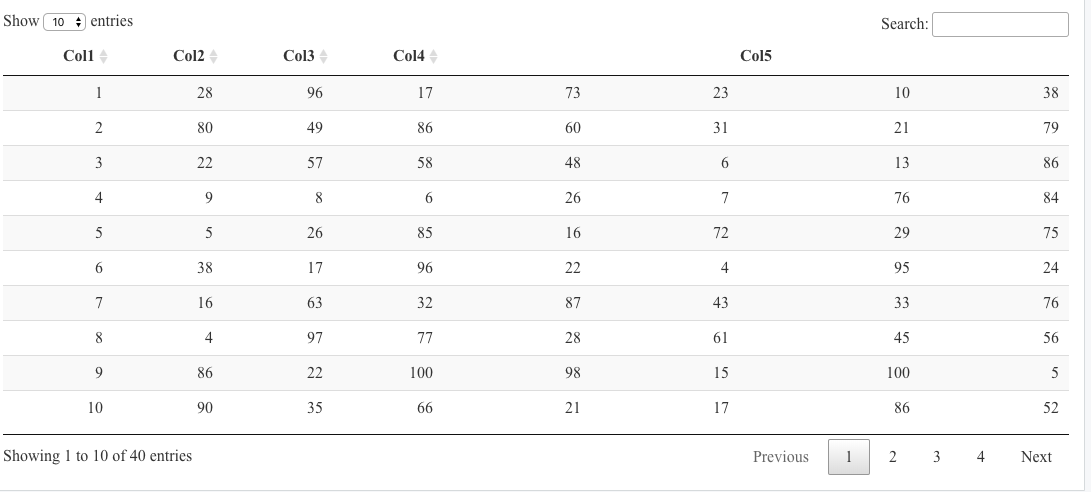
I want to group my columns in this way as shown in this 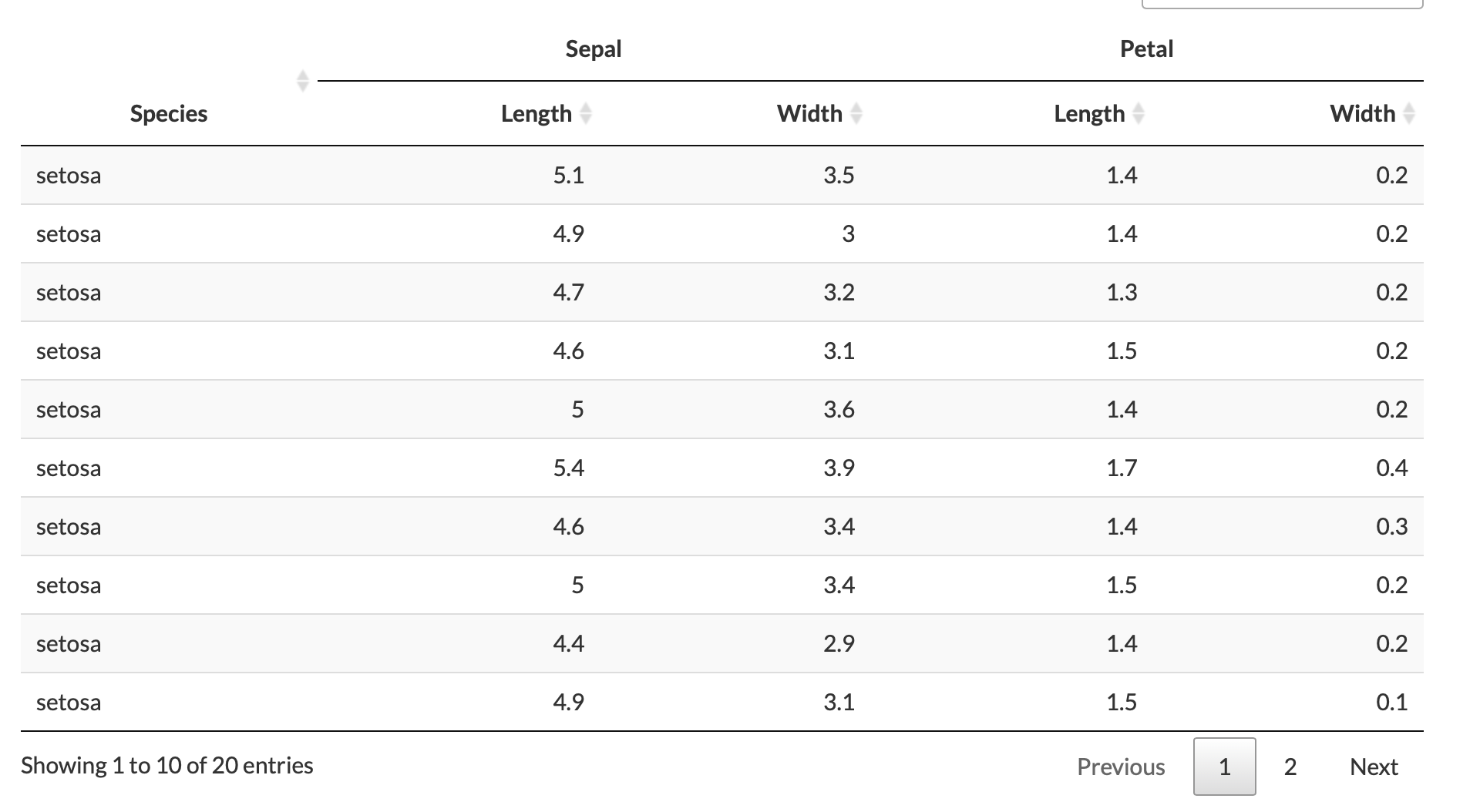
I tried this:
library(DT)
set.seed(1234)
DT <- data.frame(
"Col1" = c(1:40),
"Col2" = sample(1:100, size = 40),
"Col3" = sample(1:100, size = 40),
"Col4" = sample(1:100, size = 40),
"Col5_n" = sample(1:100, size = 40),
"Col5_25%" = sample(1:100, size = 40),
"Col5_median" = sample(1:100, size = 40),
"Col5_75%" = sample(1:100, size = 40)
)
inputPick <- list("Col5")
sketch = htmltools::withTags(table(
class = 'display',
thead(
tr(
th(rowspan = 2, "Col1"),
th(rowspan = 2, "Col2"),
th(rowspan = 2, "Col3"),
th(rowspan = 2, "Col4"),
lapply(inputPick,th,colspan=4,rowspan=1)
),
),
tr(
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
lapply(rep(c("n","25%","median","75%"),length(inputPick)),th)
)
)
)
print(sketch)
datatable(DT, container = sketch, rownames = FALSE)
This is how it looks:
As you can see, the table is empty and Col1, Col2, Col3, and Col4 should be on the same level as the subheaders (n,25%,median,75%). If I change the sketch code to rowspan = 1 for columns 1 to 4 the datatable is filled, but then I lose the subheaders for column 5:
sketch = htmltools::withTags(table(
class = 'display',
thead(
tr(
th(rowspan = 1, "Col1"),
th(rowspan = 1, "Col2"),
th(rowspan = 1, "Col3"),
th(rowspan = 1, "Col4"),
lapply(inputPick,th,colspan=4,rowspan=1)
),
),
tr(
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
lapply(rep(c("n","25%","median","75%"),length(inputPick)),th)
)
)
)
It is important to add that this will be included in a reactive function using Shiny, so the number of variables in DT and the corresponding variable names in inputPick will change. This is an example:
set.seed(1234)
DT <- data.frame(
"Col1" = c(1:40),
"Col2" = sample(1:100, size = 40),
"Col3" = sample(1:100, size = 40),
"Col4" = sample(1:100, size = 40),
"Col5_n" = sample(1:100, size = 40),
"Col5_25%" = sample(1:100, size = 40),
"Col5_median" = sample(1:100, size = 40),
"Col5_75%" = sample(1:100, size = 40),
"Col6_n" = sample(1:100, size = 40),
"Col6_25%" = sample(1:100, size = 40),
"Col6_median" = sample(1:100, size = 40),
"Col6_75%" = sample(1:100, size = 40)
)
inputPick <- list("Col5","Col6")
sketch = htmltools::withTags(table(
class = 'display',
thead(
tr(
th(rowspan = 1, "Col1"),
th(rowspan = 1, "Col2"),
th(rowspan = 1, "Col3"),
th(rowspan = 1, "Col4"),
lapply(inputPick,th,colspan=4,rowspan=1)
),
),
tr(
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
th(colspan = 1, "", rowspan=2),
lapply(rep(c("n","25%","median","75%"),length(inputPick)),th)
)
)
)
print(sketch)
datatable(DT, container = sketch, rownames = FALSE)
Having said all of that, does anyone know how to go about fixing this problem?
Thanks in advance!
CodePudding user response:
The DT post demonstrates it clearly. However, the post section was not very transparent about parameter descriptions in the container. See description below.
library(DT)
set.seed(1234)
DT <- data.frame(
"Col1" = c(1:40),
"Col2" = sample(1:100, size = 40),
"Col3" = sample(1:100, size = 40),
"Col4" = sample(1:100, size = 40),
"Col5_n" = sample(1:100, size = 40),
"Col5_25%" = sample(1:100, size = 40),
"Col5_median" = sample(1:100, size = 40),
"Col5_75%" = sample(1:100, size = 40),
"Col6_n" = sample(1:100, size = 40),
"Col6_25%" = sample(1:100, size = 40),
"Col6_median" = sample(1:100, size = 40),
"Col6_75%" = sample(1:100, size = 40)
)
sketch = htmltools::withTags(table(
class = 'display',
thead(
tr(
th(rowspan = 2, 'Col1'),
th(rowspan = 2, 'Col2'),
th(rowspan = 2, 'Col3'),
th(rowspan = 2, 'Col4'),
th(colspan = 4, 'Col5'),
th(colspan = 4, 'Col6')
),
tr(
lapply(rep(c('n', '25%', 'median', '75%'), 2), th)
)
)
))
datatable(DT, container = sketch, rownames = FALSE)
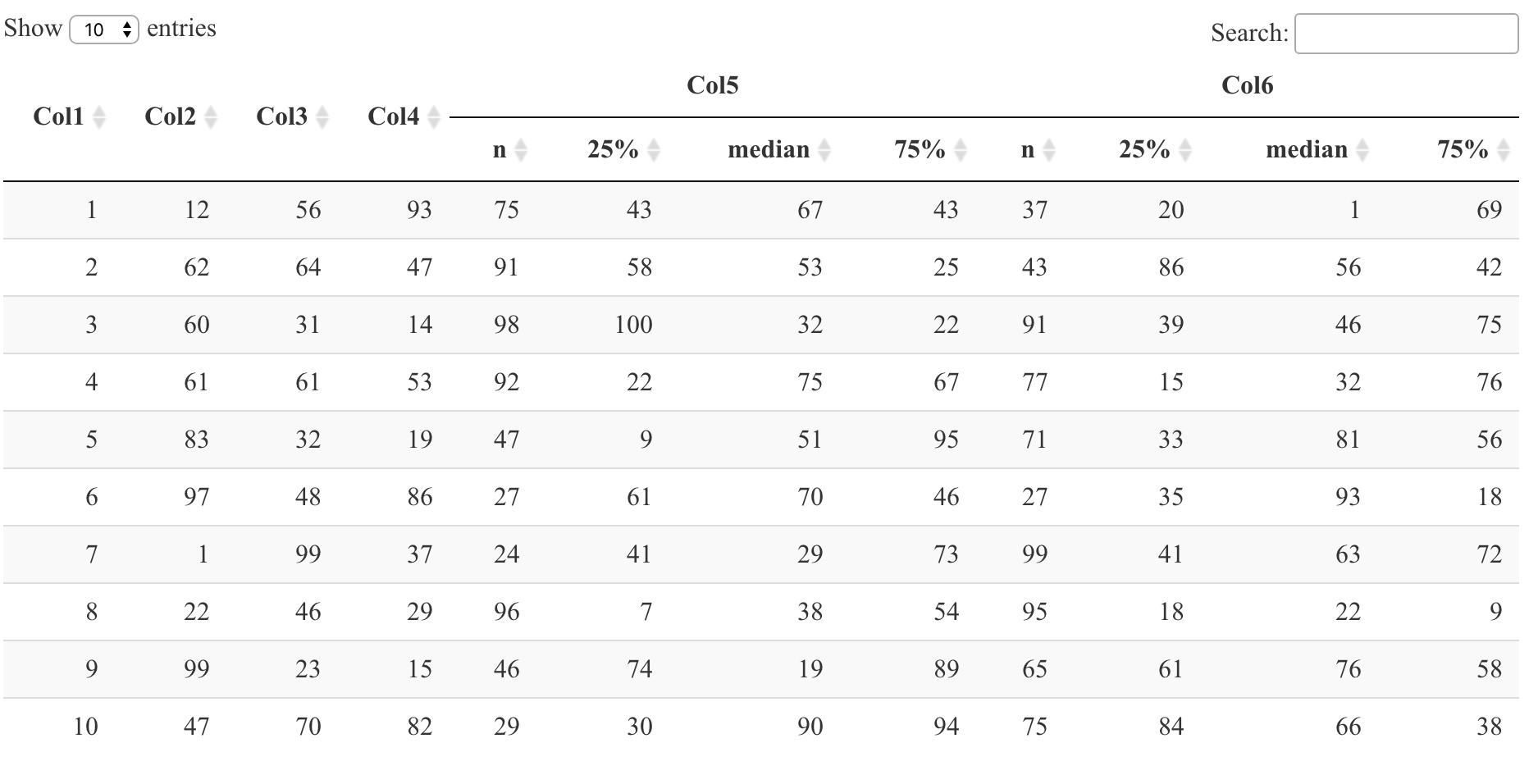
Note that within sketch,
th(rowspan = 2, 'Col1') means that "Col1" column title takes up two rows. I did the same for "Col2" through "Col4".
th(colspan = 4, 'Col5') means that "Col5" column title is overtop four columns. I did the same for "Col6".