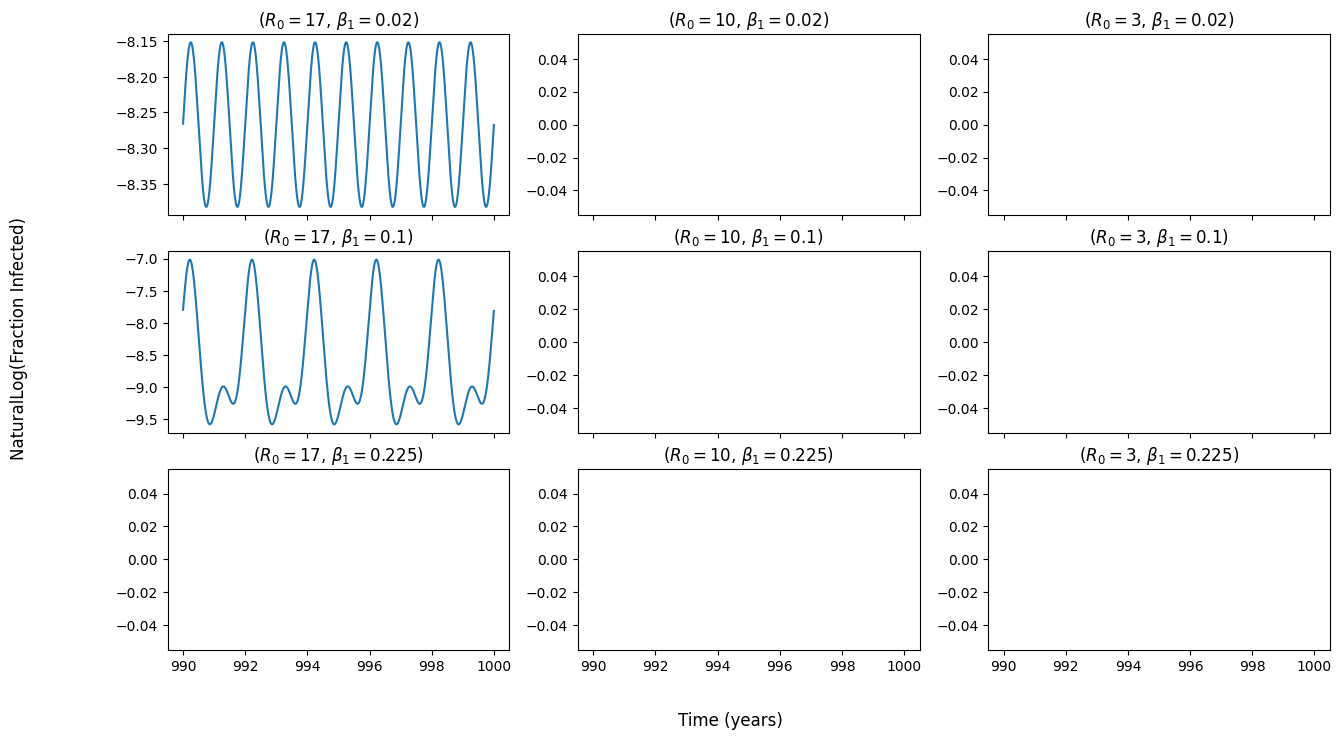
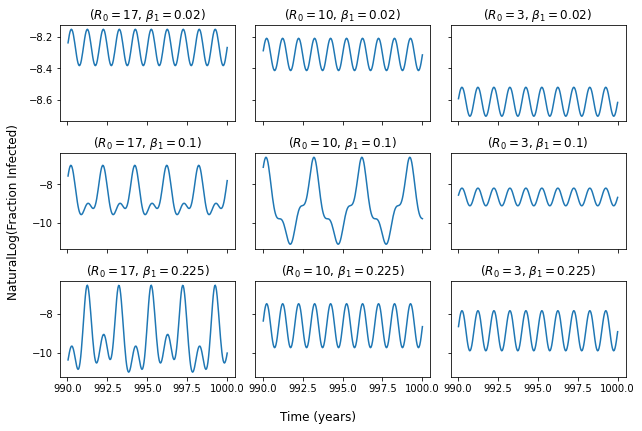
I am trying to reproduce figure 5.6 (attached) from the textbook "Modeling Infectious Diseases in Humans and Animals (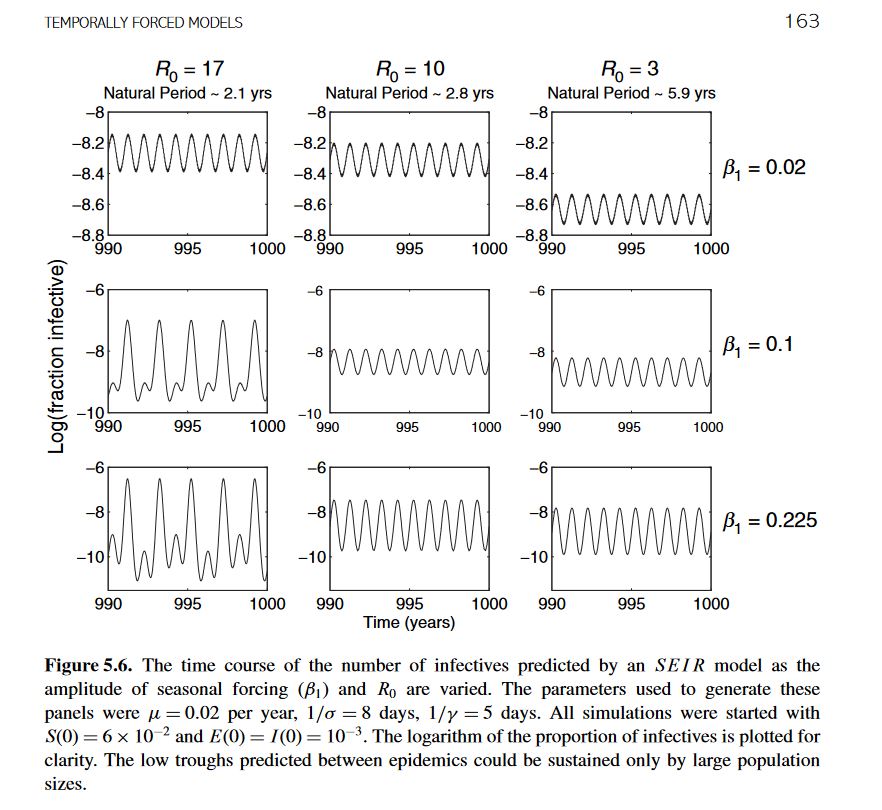
My figure within the same time range (990 - 1000 years). Natural log taken of fraction infected:
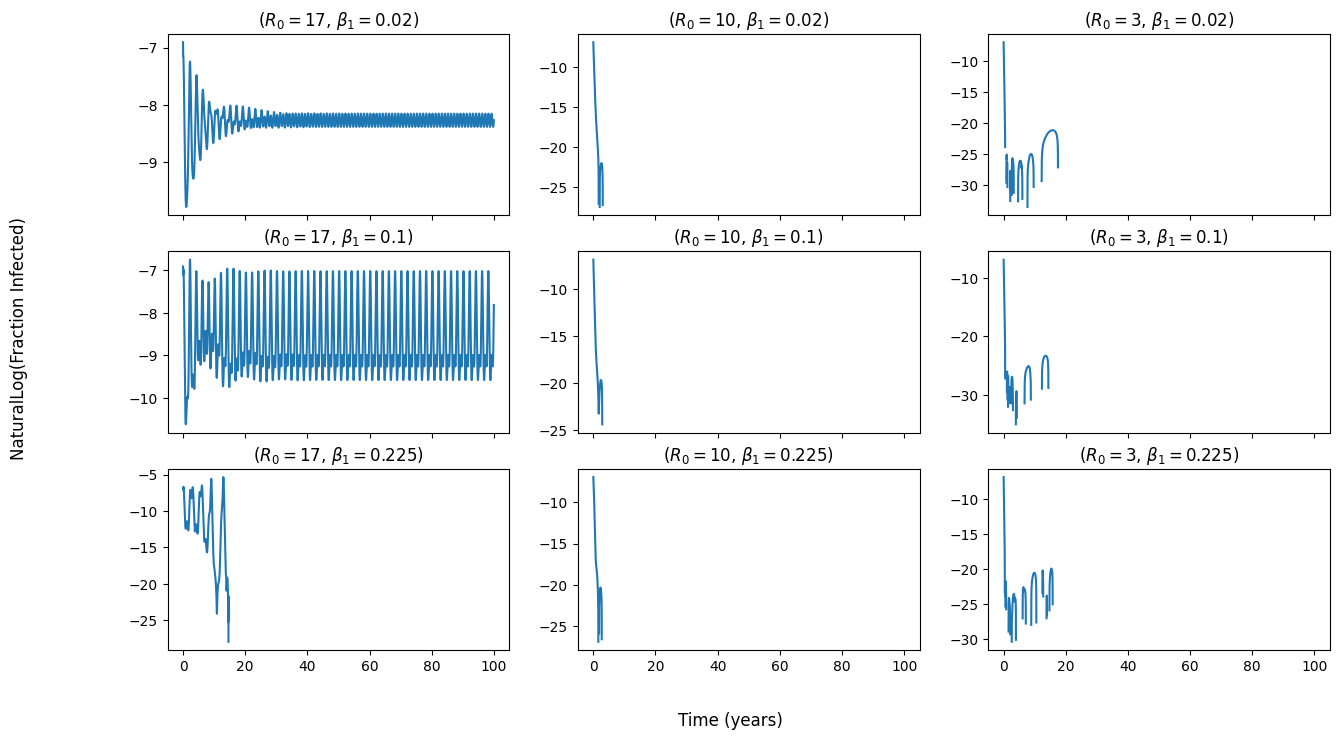
My figure but with a shorter time range (0 - 100 years). Natural log taken of fraction infected. The numerical solution for the infected population seems to fail between the 5 and 20 year mark for most of the seasonal parameters (Beta 1 and R0):
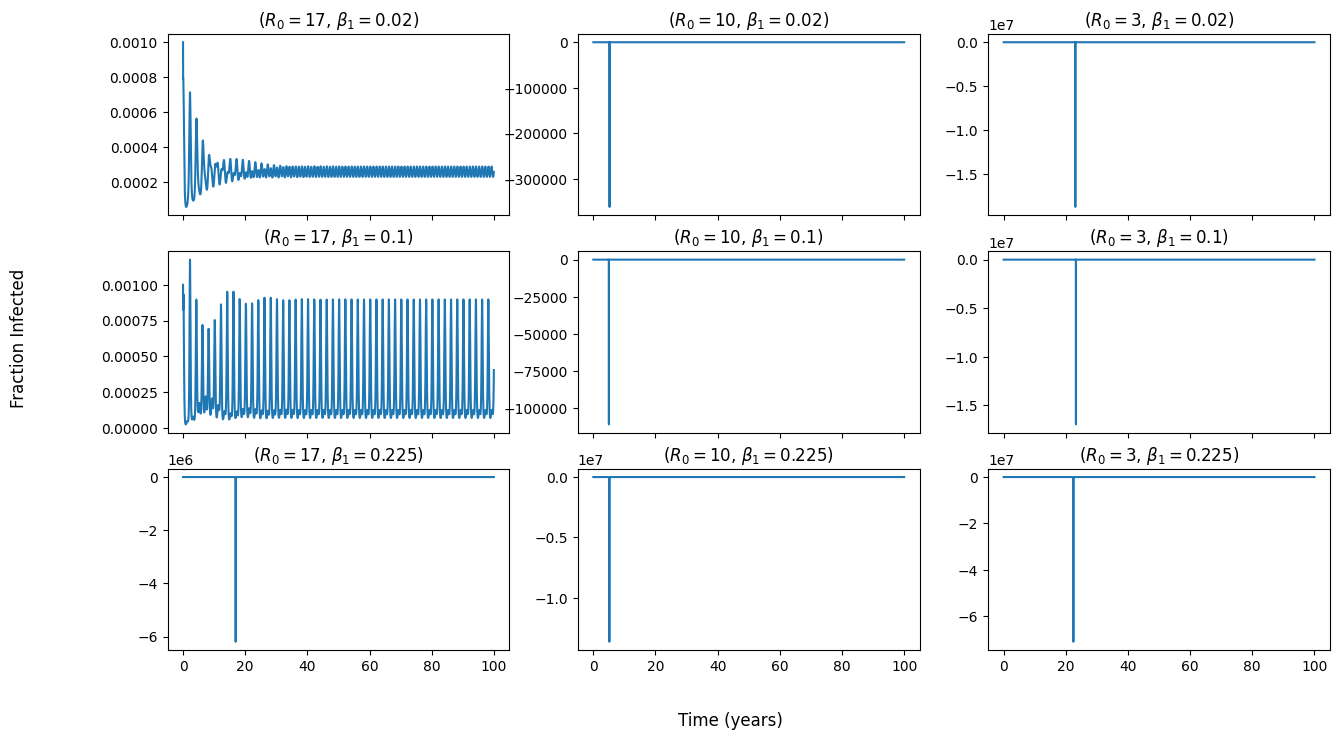
My figure with a shorter time range as above, but with no natural log taken of fraction infected.
Code to reproduce my figure:
# Code to minimally reproduce figure
import itertools
import numpy as np
import matplotlib.pyplot as plt
from scipy.integrate import odeint
def seir(y, t, mu, sigma, gamma, omega, beta_zero, beta_one):
"""System of diff eqs for epidemiological model.
SEIR stands for susceptibles, exposed, infectious, and
recovered populations.
References:
[SEIR Python Program from Textbook](http://homepages.warwick.ac.uk/~masfz/ModelingInfectiousDiseases/Chapter2/Program_2.6/Program_2_6.py)
[Seasonally Forced SIR Program from Textbook](http://homepages.warwick.ac.uk/~masfz/ModelingInfectiousDiseases/Chapter5/Program_5.1/Program_5_1.py)
"""
s, e, i = y
beta = beta_zero * (1 beta_one * np.cos(omega * t))
sdot = mu - (beta*i mu)*s
edot = beta*s*i - (mu sigma)*e
idot = sigma*e - (mu gamma)*i
return sdot, edot, idot
def solve_beta_zero(basic_reproductive_rate, gamma):
"""Defined in the last paragraph of pg. 159 of textbook Keeling 2008."""
return gamma * basic_reproductive_rate
# Model parameters (see Figure 5.6 description)
mu = 0.02 / 365
sigma = 1/8
gamma = 1/5
omega = 2 * np.pi / 365 # frequency of oscillations per year
# Seasonal forcing parameters
r0s = [17, 10, 3]
b1s = [0.02, 0.1, 0.225]
# Permutes params to get tuples matching row i column j params in figure
# e.g., [(0.02, 17), (0.02, 10) ... ]
seasonal_params = [p for p in itertools.product(*(b1s, r0s))]
# Initial Conditions: I assume these are proportions of some total population
s0 = 6e-2
e0 = i0 = 1e-3
initial_conditions = [s0, e0, i0]
# Timesteps
nyears = 1000
days_per_year = 365
ndays = nyears * days_per_year
timesteps = np.arange(1, ndays 1, 1)
# Range to slice data to reproduce my figures
# NOTE: CHange the min slice or max slice for different ranges
min_slice = 990 # or 0
max_slice = 1000 # or 100
sliced = slice(min_slice * 365, max_slice * 365)
x_ticks = timesteps[sliced]/days_per_year
# Define figure
nrows = 3
ncols = 3
fig, ax = plt.subplots(nrows, ncols, sharex=True, figsize=(15, 8))
# Iterate through parameters and recreate figure
for i in range(nrows):
for j in range(ncols):
# Get seasonal parameters for this subplot
beta_one = seasonal_params[i * nrows j][0]
basic_reproductive_rate = seasonal_params[i * nrows j][1]
# Compute beta zero given the reproductive rate
beta_zero = solve_beta_zero(
basic_reproductive_rate=basic_reproductive_rate,
gamma=gamma)
# Numerically solve the model, extract only the infected solutions,
# slice those solutions to the desired time range, and then natural
# log scale them
solutions = odeint(
seir,
initial_conditions,
timesteps,
args=(mu, sigma, gamma, omega, beta_zero, beta_one))
infected_solutions = solutions[:, 2]
log_infected = np.log(infected_solutions[sliced])
# NOTE: To inspect results without natural log, uncomment the
# below line
# log_infected = infected_solutions[sliced]
# DEBUG: For shape and parameter printing
# print(
# infected_solutions.shape, 'R0=', basic_reproductive_rate, 'B1=', beta_one)
# Plot results
ax[i,j].plot(x_ticks, log_infected)
# label subplot
ax[i,j].title.set_text(rf'$(R_0=${basic_reproductive_rate}, $\beta_{1}=${beta_one})')
fig.supylabel('NaturalLog(Fraction Infected)')
fig.supxlabel('Time (years)')
Disclaimer:
My short term solution is to simply change the list of seasonal parameters to values that will produce data for that range, and this adequately illustrates the effects of seasonal forcing. The point is to reproduce the figure, though, and if the author was able to do it, others should be able to as well.
CodePudding user response:
Your first (and possibly main) problem is one of scale. This diagnosis is also conform with the observations in your later experiments. The system is such that if it is started with positive values, it should stay within positive values. That negative values are reached is only possible if the step errors of the numerical method are too large.
As you can see in the original graphs, the range of values goes from exp(-7) ~= 9e-4 to exp(-12) ~= 6e-6. The value of the absolute tolerance should force at least 3 digits to be exact, so atol = 1e-10 or smaller. The relative tolerance should be adapted similarly. Viewing all components together shows that the first component has values around exp(-2.5) ~= 5e-2, so per-component tolerances should provide better results. The corresponding call is
solutions = odeint(
seir,
initial_conditions,
timesteps,
args=(mu, sigma, gamma, omega, beta_zero, beta_one),
atol = [1e-9,1e-13,1e-13], rtol=1e-11)
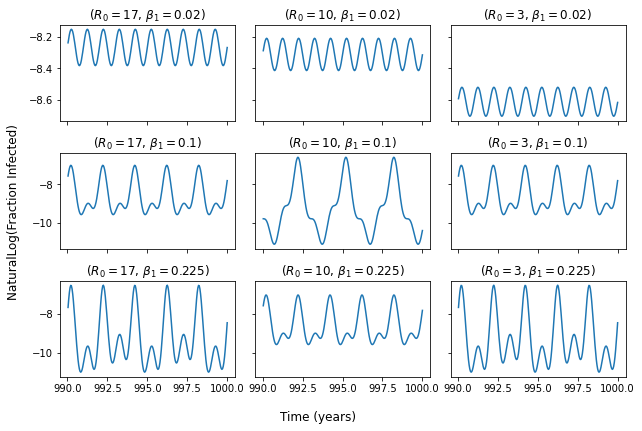
With these parameters I get the plots below
The first row and first column are as in the cited graphic, the others look different.
As a test and a general method to integrate in the range of small positive solutions, reformulate for the integration of the logarithms of the components. This can be done with a simple wrapper
def seir_log(log_y,t,*args):
y = np.exp(log_y)
dy = np.array(seir(y,t,*args))
return dy/y # = d(log(y))
Now the expected values have scale 1 to 10, so that the tolerances are no longer so critical, default tolerances should be sufficient, but it is better to work with documented tolerances.
log_solution = odeint(
seir_log,
np.log(initial_conditions),
timesteps,
args=(mu, sigma, gamma, omega, beta_zero, beta_one),
atol = 1e-8, rtol=1e-9)
log_infected = log_solution[sliced,2]
The bottom-left plot is still sensible to atol, with 1e-7 one gets a more wavy picture. Bounding the step size with hmax=5 also stabilizes that. With the code as above the plots are
The center plot is still different than the reference. It might be that there are different stable cycles.