You can use the following piece of code as an example:
options(stringsAsFactors = FALSE)
options(encoding = "UTF-8")
library(shiny)
library(shinyWidgets)
library(shinydashboard)
library(dplyr)
library(shinycssloaders)
library(data.table)
library(tidyverse)
library(DT)
dt <- structure(list(GENE = c("SI", "ARSA", "ABCA3", "KIT", "IVD", "COL18A1"), RefSeq_ID = c("NM_001041.4", "NM_000487.6", "NM_001089.3", "NM_000222.3", "NM_002225.5", "NM_001379500.1"), Tag = c("DM", "DM", "DM", "DM", "DM?", "DM"), clinvar_clnsig = c("Pathogenic", "Uncertain_significance", "NULL", "NULL", "NULL", "Likely_pathogenic"), MutationType = c("missense", "missense", "initiation", "missense", "missense", "nonsense"), ExpectedInheritance = c("AR", "AR", "AR", "AD", "AR", "AR"), Disease = c("Sucrase isomaltase deficiency", "Metachromatic leukodystrophy", "Fatal surfactant deficiency", "Piebaldism", "Isovaleric acidaemia", "Knobloch syndrome"), hgvs = c("1022T>C", "991G>A", "3G>C", "1861G>T", "1124G>A", "1876C>T"), hgvsAll = c("1022TtoC | L341P", "991GtoA | E331K", "3GtoC | M1I", "1861GtoT | A621S", "1124GtoA | G375D", "1876CtoT | R626*"), comments = c("Descr. as T/C 1021 L340P, mut. conf. by PC <1592>.", "Found in cis with Pd allele. Descr. as G985A E329R, mut. conf. by PC <1251>.", "Descr. as M1I, base change conf. by PC <1663>.", "Descr. as 1861G>C A621S, mut. conf. by PC <1495>.", "Descr. as c.1124G>A; G375A, mut. conf. by PC <1331>.", "Descr. as c.2416C>T, posn. conf. by PC <1439>."), gnomad_AC = c("NULL", "NULL", "NULL", "NULL", "NULL", "NULL"), pmid = c("10903344", "12809637", "16641205", "17124503", "19089597", "18484314"), pmidAll = c("NULL", "30052522", "24871971", "NULL", "32778825|32977617", "16532212"), CHROM = c("3", "22", "16", "4", "15", "21"), POS = c("165060026", "50626052", "2326464", "54727909", "40416348", "45487489"), REF = c("A", "C", "C", "G", "G", "C"), ALT = c("G", "T", "G", "T", "A", "T"), Support = c("NULL", "1", "0", "NULL", "2", "1"), Rankscore = c("0.48", "0.17", "0.38", "0.68", "0.5871645293736492", "0.99"), gdbid = c("120377", "119007", "3770735", "120117", "119354", "138752"), omimid = c("609845", "607574", "601615", "164920", "607036", "120328"), amino = c("Leu-Pro", "Glu-Lys", "Met-Ile", "Ala-Ser", "Gly-Asp", "Arg-Term"), deletion = c("NULL", "NULL", "NULL", "NULL", "NULL", "NULL"), insertion = c("NULL", "NULL", "NULL", "NULL", "NULL", "NULL"), codon = c("341", "331", "1", "621", "375", "626"), codonAff = c("341", "331", "1", "621", "375", "626"), descr = c("Leu341Pro", "Glu331Lys", "Met1Ile", "Ala621Ser", "Gly375Asp", "Arg626Term"), refseq = c("NM_001041.4", "NM_000487.6", "NM_001089.3", "NM_000222.3", "NM_002225.5", "NM_001379500.1"), dbsnp = c("rs267607049", "NULL", "NULL", "NULL", "rs769261274", "NULL"), gnomad_AF = c("NULL", "NULL", "NULL", "NULL", "NULL", "NULL"), gnomad_AN = c("NULL", "NULL", "NULL", "NULL", "NULL", "NULL"), new_date = c("2013-06-11", "2011-09-23", "2013-09-19", "2012-11-06", "2012-01-25", "2012-07-27"), base = c("M", "M", "M", "M", "M", "M"), clinvarID = c("1413", "556001", "NULL", "NULL", "NULL", "915432"), entrezID = c("6476", "410", "21", "3815", "3712", "80781"), hgncID = c("10856", "713", "33", "6342", "6186", "2195"), svar = c("NULL", "NULL", "NULL", "NULL", "NULL", "NULL"), mut = c("Y", "Y", "Y", "Y", "Y", "Y"), poly = c("Y", "Y", "Y", "Y", "N", "Y"), ftv = c("N", "N", "N", "N", "N", "Y"), TotalMutations = c(74L, 320L, 338L, 144L, 157L, 76L), NewMutations = c(1L, 5L, 14L, 2L, 1L, 5L), gene_date = c("1996-04-01", "1996-04-01", "2004-04-15", "1996-04-01", "1996-04-01", "2000-09-15"), author = c("Jacob", "Rafi", "Garmany", "Bondanza", "Bonilla Guerrero", "Williams"), title = c("Congenital sucrase-isomaltase deficiency arising from cleavage and secretion of a mutant form of the enzyme.", "Disease-causing mutations in cis with the common arylsulfatase A pseudodeficiency allele compound the difficulties in accurately identifying patients and carriers of metachromatic leukodystrophy.", "Surfactant composition and function in patients with ABCA3 mutations.", "Piebald trait: implication of kit mutation on in vitro melanocyte survival and on the clinical application of cultured epidermal autografts.", "Essential fatty acid profiling for routine nutritional assessment unmasks adrenoleukodystrophy in an infant with isovaleric acidaemia.", "A phenotypic variant of Knobloch syndrome."), fullname = c("J Clin Invest", "Mol Genet Metab", "Pediatr Res", "J Invest Dermatol", "J Inherit Metab Dis", "Ophthalmic Genet"), allname = c("The Journal of clinical investigation", "Molecular genetics and metabolism", "Pediatric research", "The Journal of investigative dermatology", "Journal of inherited metabolic disease", "Ophthalmic genetics"), vol = c("106", "79", "59", "127", "31S2", "29"), page = c("281", "83", "801", "676", "S453", "85"), year = c(2000L, 2003L, 2006L, 2007L, 2008L, 2008L), reftag = c("PRI", "PRI", "PRI", "PRI", "PRI", "PRI"), Some_ID = c("BM0042985", "BM0393251", "BM0673028", "BM0795183", "BM0867669", "BM0887391"), OtherNames = c("NULL", "ASA|MLD", "ABC-C|ABC3|EST111653|LBM180|SMDP3", "C-Kit|CD117|MASTC|PBT|SCFR", "ACAD2|IVDH", "GLCC|KNO|KNO1|KS"), Location = c("3q25.2-q26.2", "22q13.31-qter", "16p13.3", "4q11-q12", "15q14-q15", "21q22.3"), STRAND = c("-", "-", "-", " ", " ", " "), FullGeneName = c("Sucrase-isomaltase", "Arylsulfatase A", "ATP binding cassette subfamily A member 3", "KIT proto-oncogene, receptor tyrosine kinase", "Isovaleryl-CoA dehydrogenase", "Collagen type XVIII alpha 1 chain")), row.names = c(NA, -6L), class = c("data.table", "data.frame"))
dt$gnomad_AC <- as.numeric(dt$gnomad_AC)
dt$Support <- as.numeric(dt$Support)
dt$Rankscore <- as.numeric(dt$Rankscore)
dt$gnomad_AF <- as.numeric(dt$gnomad_AF)
dt$gnomad_AN <- as.numeric(dt$gnomad_AN)
# ui
ui <- fluidPage(
theme = "slate",
navbarPage(
title = "Some Table",
header = tagList(useShinydashboard()),
tabPanel(
"Test",
fluidRow(
box(
dataTableOutput("mytable") %>% withSpinner(color="#0dc5c1"),
width = 12,
collapsible = FALSE,
title = "",
solidHeader = T
)
)
)
)
)
# server
server <- function(input, output) {
res <- reactive ({
outputdf <- withProgress(
message = "Loading ...",
expr = {sample_n(dt, size = 5) }
)
outputdf
})
output$mytable <-
renderDataTable(
res(),
filter = list(position = "top", clear = FALSE, plain = TRUE),
options = list(scrollX = TRUE,autoWidth = TRUE, search = list(regex = TRUE)),
rownames = FALSE
)
}
# app
shinyApp(ui, server)
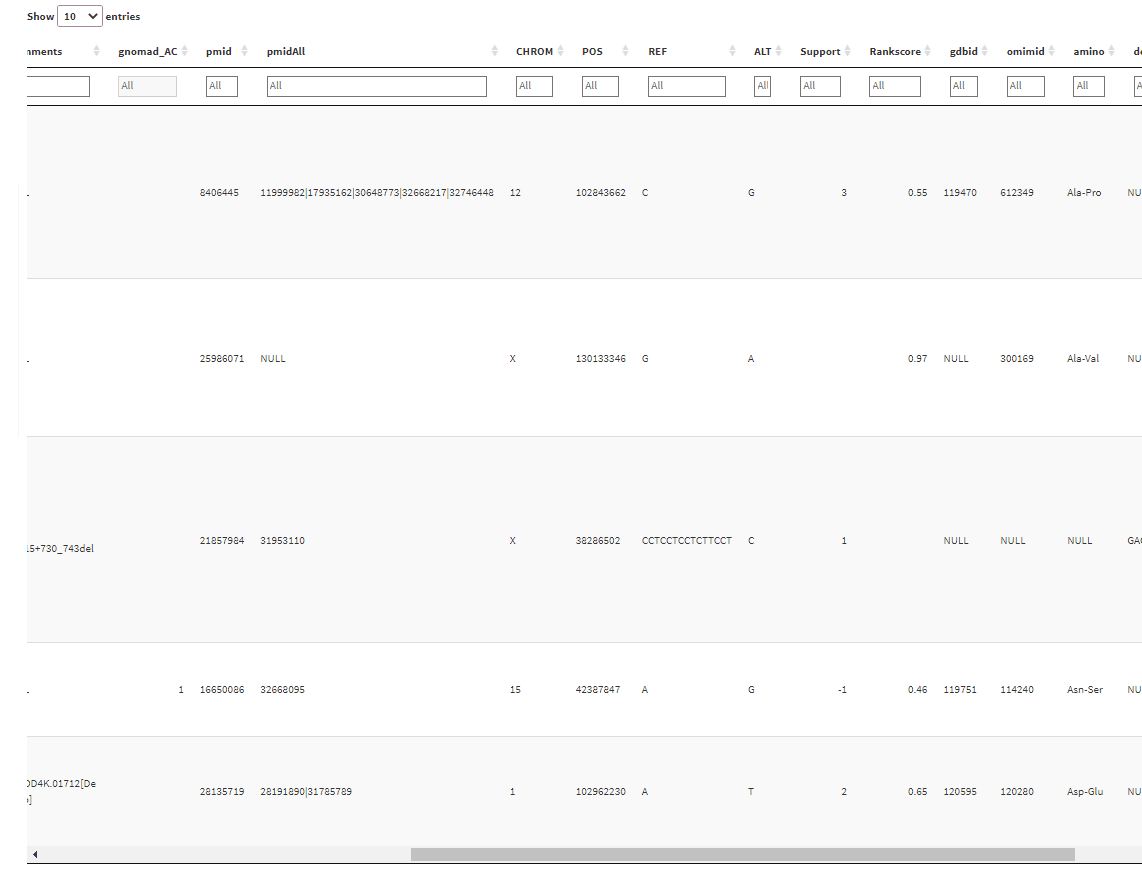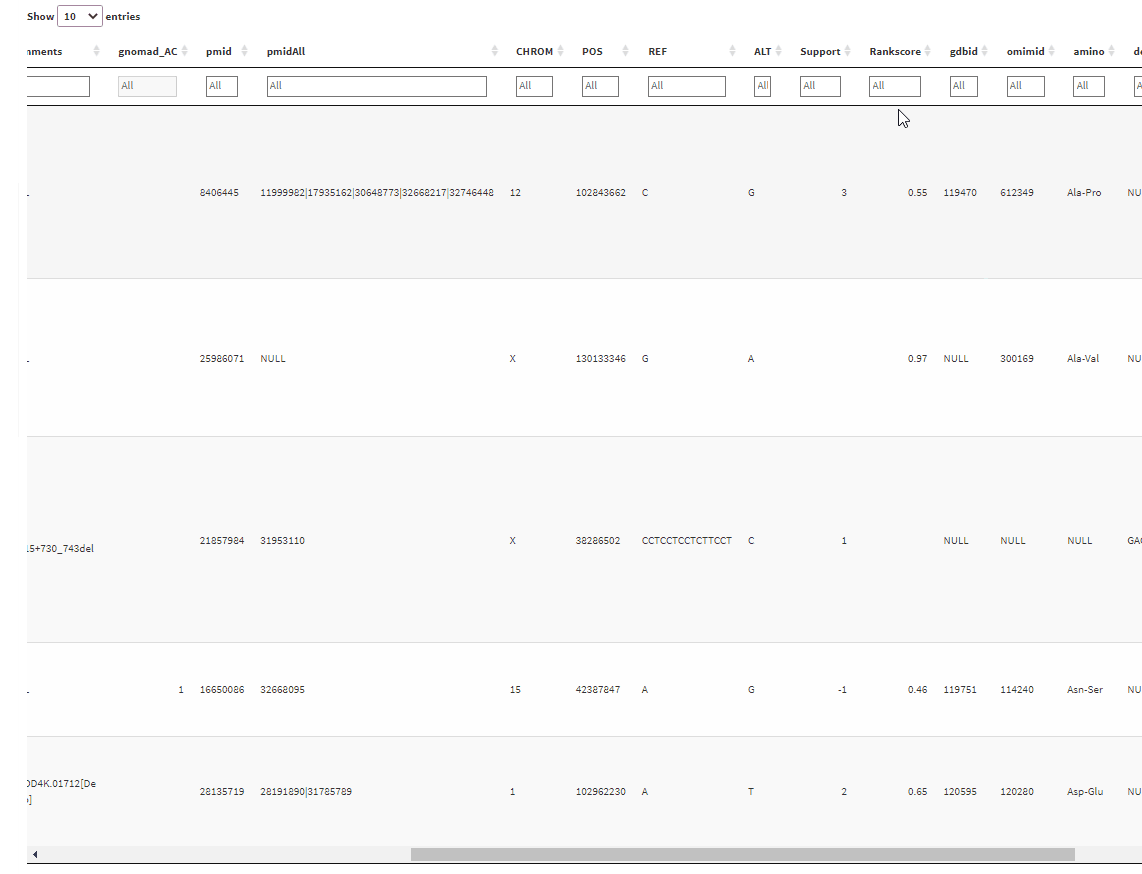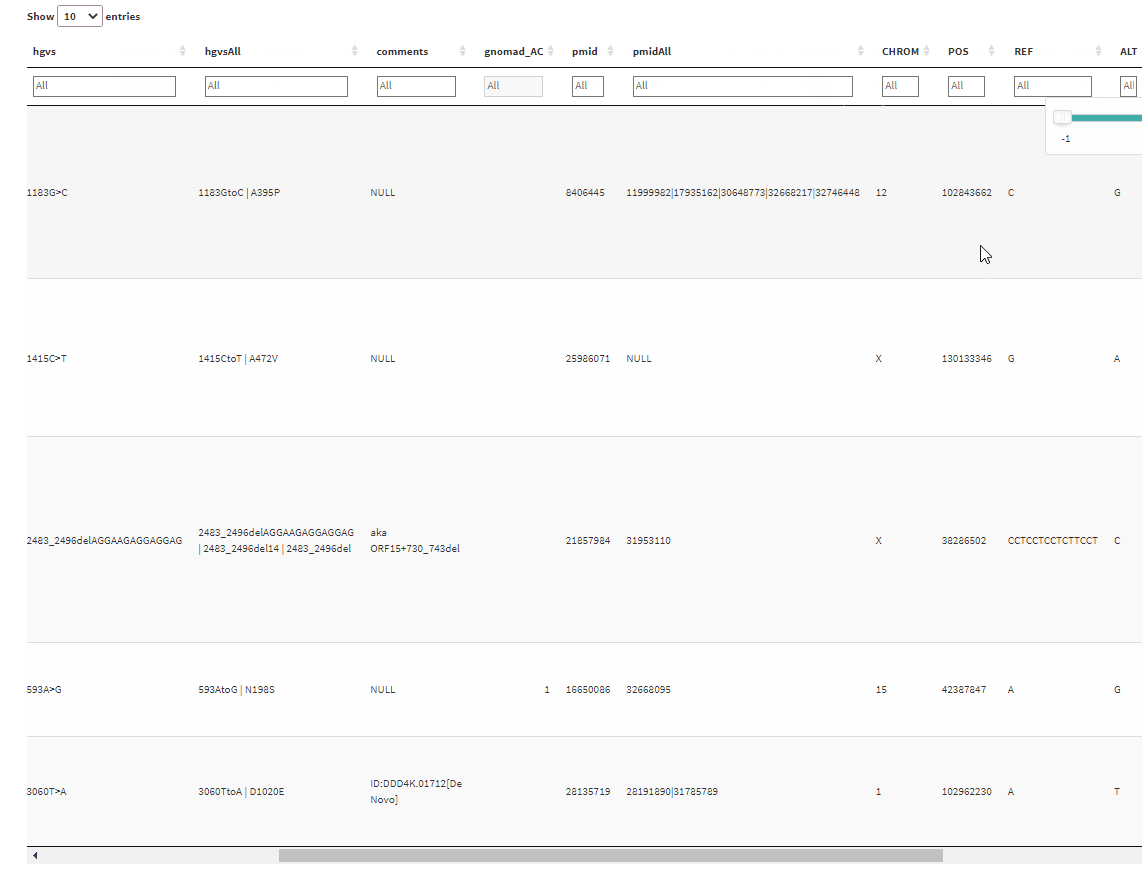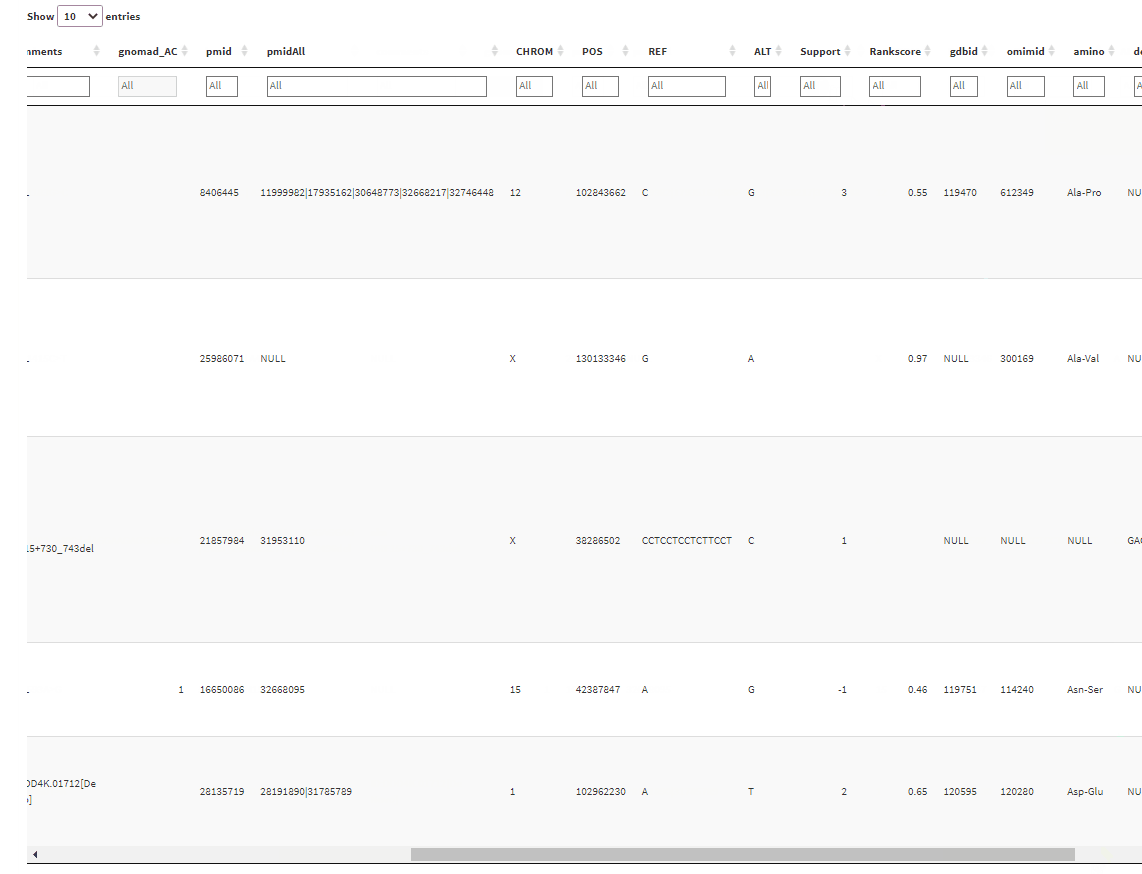
And the output will give a table with filtering boxes on top of the table:
But the problem is that whenever I click on one of the filter boxes, it sends me back to the left side of the table. I have to scroll back again to where I clicked before I can use the filtering slider. Is there any way I can fix this?
I really appreciate any help you can provide.
CodePudding user response:
This isn't a new issue. It looks like it's been reported a few times to the maintainers of the package.
In your UI, you can set the box to scroll. In the server, set your table to not scroll and not set an auto width.
The changes in the user interface.
ui <- fluidPage(
theme = "slate",
navbarPage(
title = "Some Table",
header = tagList(useShinydashboard()),
tabPanel(
"Test",
fluidRow(
box(style = "overflow-x: scroll;", # <--- I'm new!
dataTableOutput("mytable") %>% withSpinner(color = "#0dc5c1"),
width = 12,
collapsible = FALSE,
title = "",
solidHeader = T
)
)
)
)
)
The change in server.
# server
server <- function(input, output) {
res <- reactive ({
outputdf <- withProgress(
message = "Loading ...",
expr = {sample_n(dt, size = 5) }
)
outputdf
})
output$mytable <-
renderDataTable(
res(),
filter = list(position = "top", clear = FALSE, plain = TRUE),
options = list(scrollX = F, autoWidth = F, # <---- both of these have flipped
search = list(regex = TRUE)),
rownames = FALSE
)
}