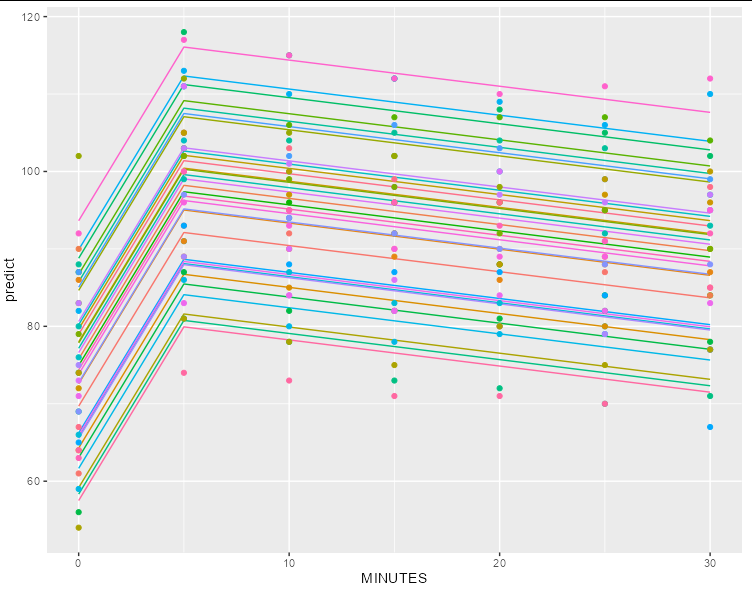
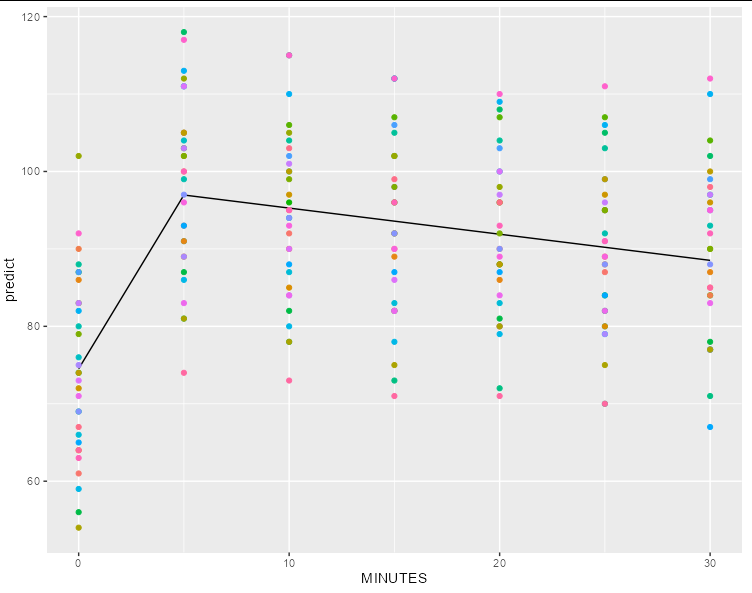
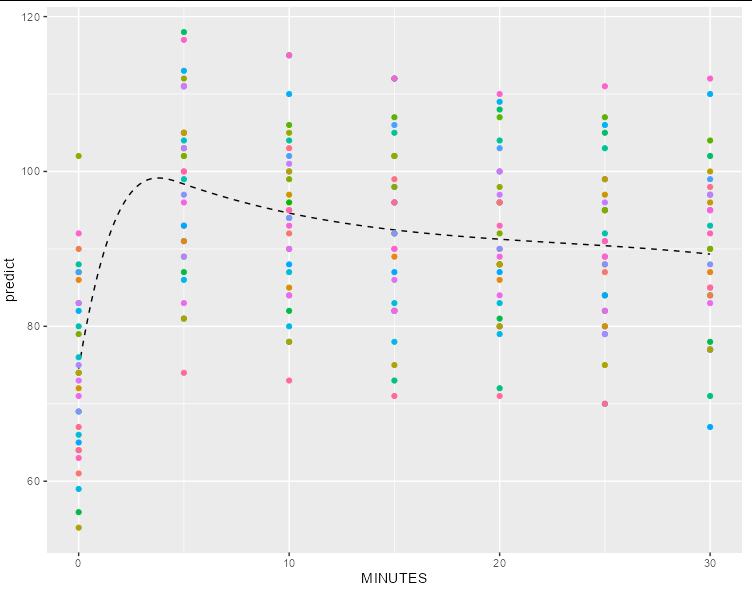
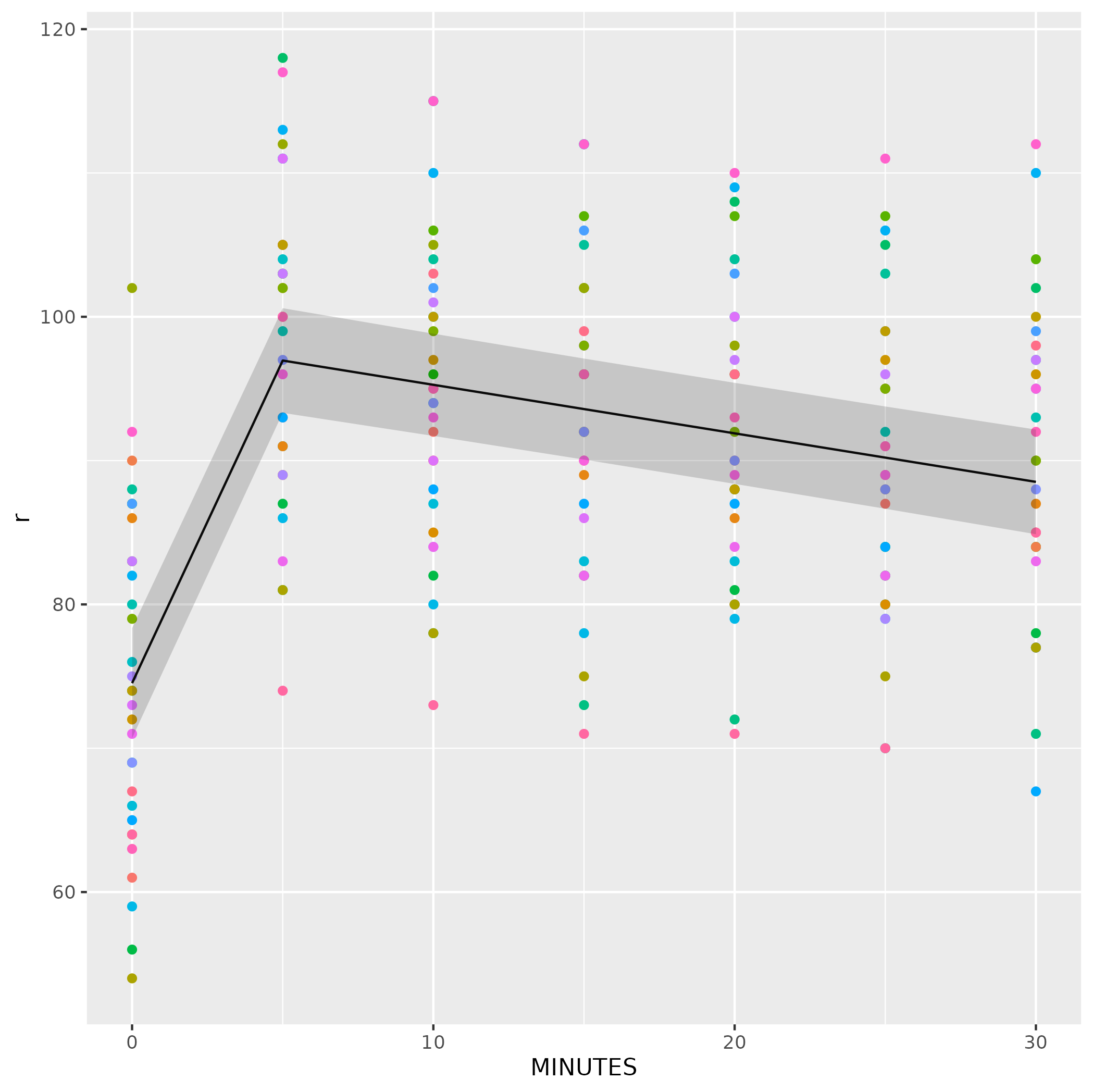
Second, without:
df$predict <- predict(m9, newdata = df, re.form = NA)
ggplot(df[1:31,], aes(MINUTES, predict, color = factor(ID)))
geom_line(color = 'black')
geom_point(data = dat, aes(y = r))
guides(color = guide_none())
If you were actually looking for a smoothed fit using splines, then you could do something like:
m9 <- lmer(r ~ splines::bs(MINUTES, knots = c(5)) ( 1 | ID) ,
data = dat )
df <- data.frame(MINUTES = rep(seq(0, 30, 0.1), length(unique(dat$ID))),
ID = rep(unique(dat$ID), each = 301))
df$predict <- predict(m9, newdata = df, re.form = NA)
ggplot(df[1:301,], aes(MINUTES, predict, color = factor(ID)))
geom_line(color = 'black', linetype = 2)
geom_point(data = dat, aes(y = r))
guides(color = guide_none())
CodePudding user response:
To get predictions with confidence intervals, you should use one of the add-on packages (effects, ggeffects, etc.) that computes confidence intervals for lmer models (note that these CIs are conditioned on the random effects estimates, i.e. they take account only of uncertainty in the fixed-effect coefficient estimates):
library(ggeffects)
pdat <- ggpredict(m9)
ggplot(dat, aes(MINUTES))
geom_point(aes(y = r, color = factor(ID)))
theme(legend.position = "none")
geom_line(data = pdat$MINUTES, aes(x = x, y = predicted))
geom_ribbon(data = pdat$MINUTES, aes(x = x, ymin = conf.low, ymax = conf.high), alpha = 0.2)