The composition of my data has 4 parts, all_data <- rbind(data1,data2,data3,data4)
In shiny,I want to when I slider sliderinput 1,only data1 changed in this dataframe.
When I change slider 2,only data2 changed in this dataframe.
But when I changed every slider,all data will change ,maybe I should use observeEvent like this,but I am faild.
observeEvent(ignoreInit = TRUE,
c(input$Mean1,input$SD1), {
mean1 <- input$Mea1
sd1 <- input$SD1
n_sample <- 5
x1 <- 1:5
y1 <- rnorm(n_sample,mean1, sd1)
z1 <- rep("Distribution 1",length(x1) )
mean_1 <- rep(mean(y1),length(x1))
data1 <-data.frame(x = x1,y = y1,z = z1,my_mean =mean_1)
})
There is all my code, could you please tell me how to modify it. Thanks!
rm(list = ls())
library(shiny)
library(shinythemes)
library(ggplot2)
library(plotly)
ui_chart <- sidebarPanel(width =3,position = "right",
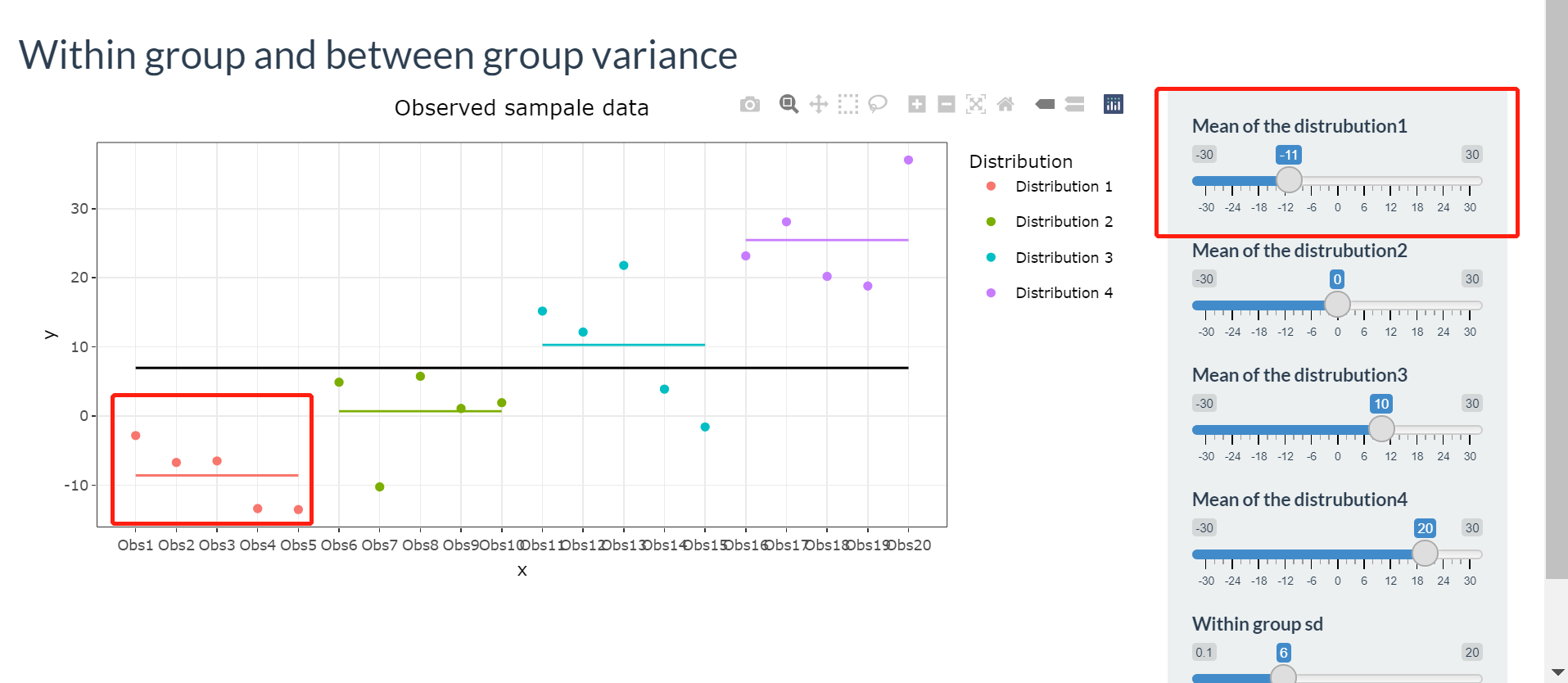
sliderInput("Mean1", "Mean of the distrubution1", -30, 30, value = -10),
sliderInput("Mean2", "Mean of the distrubution2", -30, 30, value = 0),
sliderInput("Mean3", "Mean of the distrubution3", -30, 30, value = 10),
sliderInput("Mean4", "Mean of the distrubution4", -30, 30, value = 20),
sliderInput("SD1", "Within group sd", 0.1, 20, value =6)
)
ui <- navbarPage("Distrubution for each group", theme = shinytheme("flatly"),
tabPanel("Distrubution simulation",
titlePanel("Within group and between group variance"),
mainPanel(width = 9
,plotlyOutput("chart2")
)
,ui_chart
)
)
server <- function(input, output) {
output$chart2 <- renderPlotly({
mean1 <- input$Mean1
mean2 <- input$Mean2
mean3 <- input$Mean3
mean4 <- input$Mean4
sd1 <- input$SD1
n_sample <- 5
x1 <- 1:5
y1 <- rnorm(n_sample,mean1, sd1)
z1 <- rep("Distribution 1",length(x1) )
mean_1 <- rep(mean(y1),length(x1))
data1 <-data.frame(x = x1,y = y1,z = z1,my_mean =mean_1 )
x2 <- 6:10
y2 <- rnorm(n_sample,mean2, sd1)
z2 <- rep("Distribution 2",length(x2) )
mean_2 <- rep(mean(y2),length(x2))
data2 <-data.frame(x = x2,y = y2,z = z2,my_mean =mean_2)
x3 <- 11:15
y3 <- rnorm(n_sample,mean3, sd1)
z3 <- rep("Distribution 3",length(x3) )
mean_3 <- rep(mean(y3),length(x3))
data3 <-data.frame(x = x3,y = y3,z = z3,my_mean =mean_3)
x4 <- 16:20
y4 <- rnorm(n_sample,mean4, sd1)
z4 <- rep("Distribution 4",length(x4) )
mean_4 <- rep(mean(y4),length(x4))
data4 <-data.frame(x = x4,y = y4,z = z4,my_mean =mean_4)
all_data <- rbind(data1,data2,data3,data4)
all_data$mean_all <- mean(all_data$y)
p2 <- ggplot(data = all_data)
geom_point(aes(x = x ,y = y,color = z))
geom_line(aes(x = x ,y = my_mean ,color = z))
geom_line(aes(x = x ,y = mean_all ),color = "black")
scale_x_continuous(breaks = c(1:20),
labels = paste0("Obs",1:20))
ggtitle("Observed sampale data")
theme_bw()
labs(color = "Distribution")
theme(plot.title = element_text(hjust = 0.5))
ggplotly(p2)
})
}
shinyApp(ui, server)
CodePudding user response:
Well since you are using rnorm, you are getting new random values every-time an input changes. If you want to isolate those changes, you might want to make separate reactive data objects for each of the studies. Here's one way to do that
server <- function(input, output) {
generate_data <- function(dist, ids, n_sample, sample_mean, sample_sd) {
x <- 1:5
y <- rnorm(n_sample,sample_mean, sample_sd)
z <- rep(paste("Distribution", dist),length(x) )
mean <- rep(mean(y),length(x))
data.frame(x = ids, y = y, z = z, my_mean =mean)
}
n_sample <- 5
data1 <- reactive(generate_data("1", 1:5, n_sample, input$Mean1, input$SD1))
data2 <- reactive(generate_data("2", 6:10, n_sample, input$Mean2, input$SD1))
data3 <- reactive(generate_data("3", 11:15, n_sample, input$Mean3, input$SD1))
data4 <- reactive(generate_data("4", 16:20, n_sample, input$Mean4, input$SD1))
output$chart2 <- renderPlotly({
all_data <- rbind(data1(),data2(),data3(),data4())
all_data$mean_all <- mean(all_data$y)
p2 <- ggplot(data = all_data)
geom_point(aes(x = x ,y = y,color = z))
geom_line(aes(x = x ,y = my_mean ,color = z))
geom_line(aes(x = x ,y = mean_all ),color = "black")
scale_x_continuous(breaks = c(1:20),
labels = paste0("Obs",1:20))
ggtitle("Observed sampale data")
theme_bw()
labs(color = "Distribution")
theme(plot.title = element_text(hjust = 0.5))
ggplotly(p2)
})
}
Here we make a helper function to generate the data for each of the groups. We store the data in a reactive object that only depends on the group mean. Therefore they won't change when other inputs change