I am running some statistics, and need to display my results with mean separation letters over each bar. However, I am having a problem separating my mean separation indicator letters out over each bar.
Here is my data. Running this code should generate a data frame:
data <- data.frame(group = c(1,
1,
1,
1,
1,
1,
1,
1,
1,
1,
1,
1,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
3,
3,
3,
3,
3,
3,
3,
3,
3,
3,
3,
3,
4,
4,
4,
4,
4,
4,
4,
4,
5,
5,
5,
5,
5,
5,
5,
5,
5,
5,
5,
5),
habitat = c('woodlands',
'woodlands',
'woodlands',
'woodlands',
'beach',
'beach',
'beach',
'beach',
'prairie',
'prairie',
'prairie',
'prairie',
'woodlands',
'woodlands',
'woodlands',
'woodlands',
'beach',
'beach',
'beach',
'beach',
'prairie',
'prairie',
'prairie',
'prairie',
'woodlands',
'woodlands',
'woodlands',
'woodlands',
'prairie',
'prairie',
'prairie',
'prairie',
'beach',
'beach',
'beach',
'beach',
'prairie',
'prairie',
'prairie',
'prairie',
'beach',
'beach',
'beach',
'beach',
'beach',
'beach',
'beach',
'beach',
'prairie',
'prairie',
'prairie',
'prairie',
'woodlands',
'woodlands',
'woodlands',
'woodlands'),
species = c('skunk',
'fox',
'raccoon',
'gopher',
'raccoon',
'fox',
'skunk',
'gopher',
'skunk',
'fox',
'gopher',
'raccoon',
'gopher',
'skunk',
'raccoon',
'fox',
'raccoon',
'skunk',
'fox',
'gopher',
'gopher',
'fox',
'skunk',
'raccoon',
'raccoon',
'skunk',
'fox',
'gopher',
'gopher',
'fox',
'skunk',
'raccoon',
'gopher',
'skunk',
'raccoon',
'fox',
'skunk',
'fox',
'raccoon',
'gopher',
'skunk',
'gopher',
'raccoon',
'fox',
'skunk',
'gopher',
'fox',
'raccoon',
'skunk',
'gopher',
'raccoon',
'fox',
'fox',
'raccoon',
'gopher',
'skunk'),
response = c(321684201,
321684221,
321684211,
321684216,
321684201,
321684221,
321684216,
321684216,
321684211,
321684221,
321684226,
321684206,
321684216,
321684216,
321684216,
321684216,
321684211,
321684211,
321684226,
321684216,
321684221,
321684216,
321684216,
321684216,
321684211,
321684216,
321684226,
321684216,
321684216,
321684221,
321684201,
321684206,
321684216,
321684221,
321684211,
321684221,
321684206,
321684226,
321684226,
321684226,
321684211,
321684216,
321684206,
321684206,
321684211,
321684221,
321684216,
321684206,
321684171,
321684186,
321684181,
321684161,
321684226,
321684216,
321684221,
321684201))
Here is what I have tried:
# Open packages
library(lme4)
library(ggplot2)
library(emmeans)
library(car)
# Convert columns to factors
data$group <- as.factor(data$group)
data$habitat <- as.factor(data$habitat)
data$species <- as.factor(data$species)
# Generate model and investigate significance
mod <- lmer(response ~ habitat * species (1|group) (1|group:habitat), data = data)
Anova(mod, type = ("II"))
# Interaction term isn't significant at the 0.05 level, but let's assume it is for this question.
# Separate means using the emmeans package
mod_means_cotr <- emmeans(mod, pairwise ~ 'habitat:species',
adjust = 'tukey')
mod_means <- multcomp::cld(object = mod_means_cotr$emmeans,
LETTERS = "letters")
mod_means$hsd <- c('bb',
'abbb',
'abbb',
'abbb',
'abbb',
'abbb',
'abbb',
'aa',
'abbb',
'abbb',
'abbb',
'abbb') # This is done to illustrate situations with long mean separator sequences
mod_means
# Generate the graph
ggplot(mod_means, aes(fill = species, x = habitat, y = emmean))
geom_bar(stat="identity", width = 0.6, position = "dodge", col = "black")
geom_errorbar(aes(ymin = emmean, ymax = emmean SE), width = 0.3, position = position_dodge(0.6))
guides(fill = guide_legend("Species"))
xlab("Habitat") ylab("Response")
theme(plot.title=element_text(hjust=0.5, size = 25),
axis.text = element_text(size = 20),
axis.title = element_text(size = 20))
ggtitle("Graph Title")
geom_text(aes(label=hsd, y=emmean SE), vjust = -0.9, size = 5)
ylim(0, 350000000)
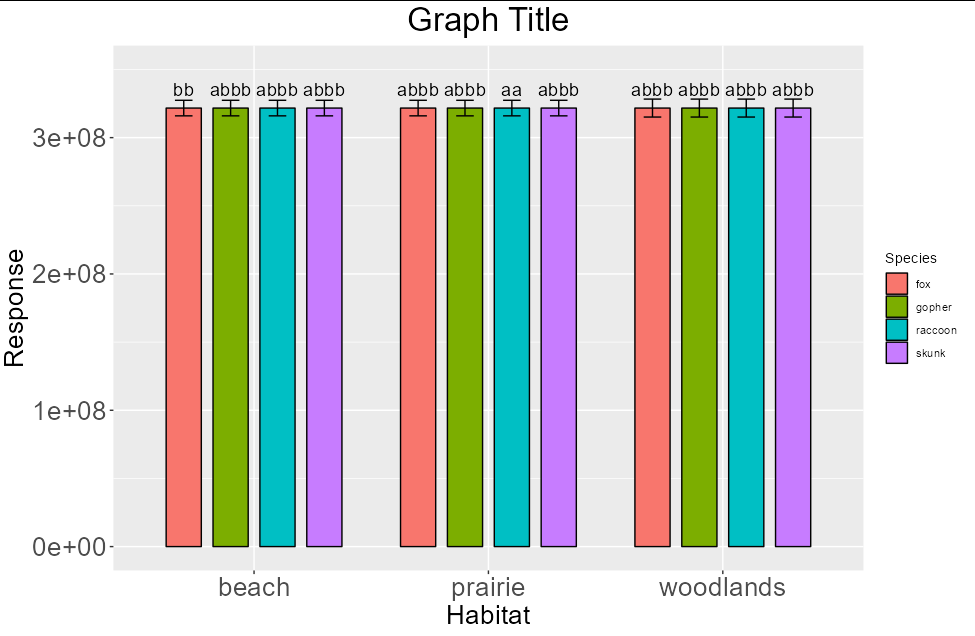
Looking at the graph, the mean separations are all overlapping and not distinguishable. In addition, the bars are not separated by any spaces. I would like to add spaces between the bars and wrap the long mean separators over each corresponding bar; I'm assuming the stringr package would come in handy, but I'm not sure. How can I do these things?
CodePudding user response:
The error bars in your example don't show up because the SE is infintesimal compared to the bars themselves, so for the purposes of example I have multiplied them by 1,000,000. Essentially the problem seems to be that you haven't used species as a grouping variable in the text or error bar layers:
# Generate the graph
ggplot(mod_means, aes(fill = species, x = habitat, y = emmean))
geom_bar(stat="identity", width = 0.6, position = position_dodge(width = 0.8),
col = "black")
geom_errorbar(aes(ymin = emmean - SE * 1e6, ymax = emmean SE * 1e6,
group = species), width = 0.3,
position = position_dodge(0.8))
guides(fill = guide_legend("Species"))
xlab("Habitat") ylab("Response")
theme(plot.title=element_text(hjust = 0.5, size = 25),
axis.text = element_text(size = 20),
axis.title = element_text(size = 20))
ggtitle("Graph Title")
geom_text(aes(label = hsd, y = emmean SE, group = species), vjust = -0.9,
size = 5, position = position_dodge(width = 0.8))
ylim(0, 350000000)