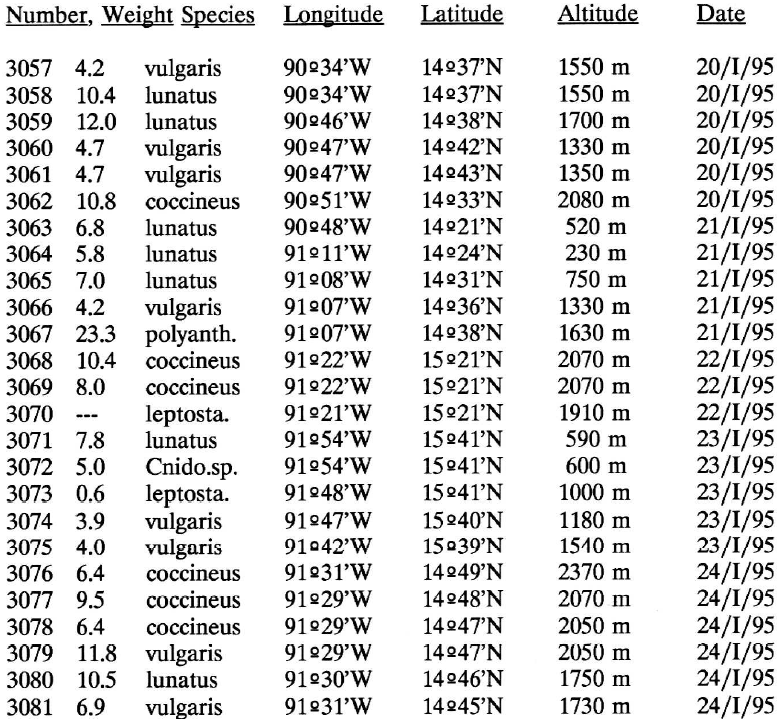
I run the code below to get the data of this picture but I encounter an error when try to set the column name.
library(magick)
library(tesseract)
team_img <- image_read("C:\\Users\\macosta\\Downloads\\Capture1.PNG") %>%
image_resize('2000x') %>%
image_convert(type = 'Grayscale') %>%
image_trim(fuzz = 40) %>%
image_write(format = 'png', density = '300x300') %>%
tesseract::ocr() %>%
strsplit('\n') %>%
getElement(1) %>%
`[`(-1) %>%
{read.table(text = .)} %>% # this part show an error
setNames(c('Number', 'Weight', 'Specie', 'long', 'lat', 'altitude', 'Date', 'Others'))
> Error in scan(file = file, what = what, sep = sep, quote = quote, dec = dec, : > line 2 did not have 9 elements
<- team_img
# [1] ""
# [2] "3057 4.2 vulgaris 90234\" W 14237N 1550 m 20/1/95"
# [3] "3058 10.4 lunatus 90234°W 14237N 1550 m 20/1/95"
# [4] "3059 12.0 lunatus 90246’W 14238’N 1700 m 20/1/95"
# [5] "3060 4.7 ~~ vulgaris 90247'W 14242’°N 1330 m 20/1/95"
# [6] "3061 4.7 vulgaris 90247 W 14243’N 1350 m 20/1/95"
# [7] "3062 10.8 coccineus 90951,W 14933N 2080m 20/1/95"
# [8] "3063 6.8 __lunatus 90248°W —-14221’N 520 m 21/1/95"
# [9] "3064 5.8 lunatus 91211\"W 14°24°N 230 m 21/1/95"
# [10] "3065 7.0 lunatus 91208’W 14231’N 750 m 21/1/95"
# [11] "3066 4.2 vulgaris 91207 W 14236°N 1330 m 21/1/95"
# [12] "3067 23.3 polyanth. 91207°W 14238’N 1630 m 21/1/95"
# [13] "3068 10.4 coccineus 91222’W 15221’N 2070 m 22/1/95"
# [14] "3069 8.0 coccineus 91222’W 15221’N 2070 m 22/1/95"
# [15] "3070. --- leptosta. 91221’W 15221’N 1910 m 22/1/95"
# [16] "3071 7.8 lunatus 91254°W 15241’N 590 m 23/1/95"
# [17] "3072 5.0 Cnido.sp. 91254°W 15241’N 600 m 23/1/95"
# [18] "3073 0.6 _leptosta. 91248°W 15241’N 1000 m 23/1/95"
# [19] "3074 3.9 vulgaris 91247'W 15240°N 1180 m 23/1/95"
# [20] "3075 4.0 vulgaris 91942\"W 15939°N 1540 m 23/1/95"
# [21] "3076 6.4 coccineus 91231’W 14249’°N 2370 m 24/1/95"
# [22] "3077 9.5 coccineus 91229°W 14248°N 2070 m 24/1/95"
# [23] "3078 64 coccineus 91229°W 14247N 2050 m 24/1/95"
# [24] "3079 11.8 vulgaris 91229’W 14247'N 2050 m 24/1/95"
# [25] "3080 10.5 lunatus 91230°W 14246’N 1750 m 24/1/95"
# [26] "3081 6.9 vulgaris 91231’°W 14245’N 1730 m 24/1/95"
Data picture.
The idea is to obtain in a DF.
CodePudding user response:
While the Tesseract OCR does a pretty admirable job of extracting text from the sample image, it seems to struggle with consistently decoding the ° and ’ symbols in the Latitude and Longitude columns. There is also a space between the measure and the units in the Altitude column. This results in strings that are interpreted by read.table as having either 7, 8 or 9 columns. Since your first input line appears to have 9 columns, read.table expects all subsequent lines to have 9 columns as well. You will have to figure out a way to clean the latitude, longitude output data so that you have 8 space-delimited columns you have identified in setNames
library(magick)
library(tesseract)
team_str <- image_read("C:\\Users\\macosta\\Downloads\\Capture1.PNG") %>%
image_resize('2000x') %>%
image_convert(type = 'Grayscale') %>%
image_trim(fuzz = 40) %>%
image_write(format = 'png', density = '300x300') %>%
tesseract::ocr() %>%
strsplit('\n') %>%
getElement(1)
sapply(team_str, strsplit, '\\s ') %>% lapply(length)
$`Number, Weight Species Longitude Latitude Altitude Date`
[1] 7
[[2]]
[1] 0
$`3057 4.2 vulgaris 90234" W 14237N 1550 m 20/1/95`
[1] 9
$`3058 10.4 lunatus 90234°W 14237N 1550 m 20/1/95`
[1] 8
$`3059 12.0 lunatus 90246’W 14238’N 1700 m 20/1/95`
[1] 8
$`3060 4.7 ~~ vulgaris 90247'W 14242’°N 1330 m 20/1/95`
[1] 9
$`3061 4.7 vulgaris 90247 W 14243’N 1350 m 20/1/95`
[1] 9
$`3062 10.8 coccineus 90951,W 14933N 2080m 20/1/95`
[1] 7
$`3063 6.8 __lunatus 90248°W —-14221’N 520 m 21/1/95`
[1] 8
$`3064 5.8 lunatus 91211"W 14°24°N 230 m 21/1/95`
[1] 8
$`3065 7.0 lunatus 91208’W 14231’N 750 m 21/1/95`
[1] 8
$`3066 4.2 vulgaris 91207 W 14236°N 1330 m 21/1/95`
[1] 9
$`3067 23.3 polyanth. 91207°W 14238’N 1630 m 21/1/95`
[1] 8
$`3068 10.4 coccineus 91222’W 15221’N 2070 m 22/1/95`
[1] 8
$`3069 8.0 coccineus 91222’W 15221’N 2070 m 22/1/95`
[1] 8
$`3070. --- leptosta. 91221’W 15221’N 1910 m 22/1/95`
[1] 8
$`3071 7.8 lunatus 91254°W 15241’N 590 m 23/1/95`
[1] 8
$`3072 5.0 Cnido.sp. 91254°W 15241’N 600 m 23/1/95`
[1] 8
$`3073 0.6 _leptosta. 91248°W 15241’N 1000 m 23/1/95`
[1] 8
$`3074 3.9 vulgaris 91247'W 15240°N 1180 m 23/1/95`
[1] 8
$`3075 4.0 vulgaris 91942"W 15939°N 1540 m 23/1/95`
[1] 8
$`3076 6.4 coccineus 91231’W 14249’°N 2370 m 24/1/95`
[1] 8
$`3077 9.5 coccineus 91229°W 14248°N 2070 m 24/1/95`
[1] 8
$`3078 64 coccineus 91229°W 14247N 2050 m 24/1/95`
[1] 8
$`3079 11.8 vulgaris 91229’W 14247'N 2050 m 24/1/95`
[1] 8
$`3080 10.5 lunatus 91230°W 14246’N 1750 m 24/1/95`
[1] 8
$`3081 6.9 vulgaris 91231’°W 14245’N 1730 m 24/1/95`
[1] 8
Combining everything with data.table::rbindlist makes it easier to visualize what is happening and to devise a strategy to deal with malformed rows:
sapply(team_str, str_split, '\\s ') %>%
lapply(t) %>%
lapply(as.data.frame) %>%
data.table::rbindlist(fill = TRUE) %>%
print
V1 V2 V3 V4 V5 V6 V7 V8 V9
1: Number, Weight Species Longitude Latitude Altitude Date <NA> <NA>
2: <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
3: 3057 4.2 vulgaris 90234" W 14237N 1550 m 20/1/95
4: 3058 10.4 lunatus 90234°W 14237N 1550 m 20/1/95 <NA>
5: 3059 12.0 lunatus 90246’W 14238’N 1700 m 20/1/95 <NA>
6: 3060 4.7 ~~ vulgaris 90247'W 14242’°N 1330 m 20/1/95
7: 3061 4.7 vulgaris 90247 W 14243’N 1350 m 20/1/95
8: 3062 10.8 coccineus 90951,W 14933N 2080m 20/1/95 <NA> <NA>
9: 3063 6.8 __lunatus 90248°W —-14221’N 520 m 21/1/95 <NA>
10: 3064 5.8 lunatus 91211"W 14°24°N 230 m 21/1/95 <NA>
11: 3065 7.0 lunatus 91208’W 14231’N 750 m 21/1/95 <NA>
12: 3066 4.2 vulgaris 91207 W 14236°N 1330 m 21/1/95
13: 3067 23.3 polyanth. 91207°W 14238’N 1630 m 21/1/95 <NA>
14: 3068 10.4 coccineus 91222’W 15221’N 2070 m 22/1/95 <NA>
15: 3069 8.0 coccineus 91222’W 15221’N 2070 m 22/1/95 <NA>
16: 3070. --- leptosta. 91221’W 15221’N 1910 m 22/1/95 <NA>
17: 3071 7.8 lunatus 91254°W 15241’N 590 m 23/1/95 <NA>
18: 3072 5.0 Cnido.sp. 91254°W 15241’N 600 m 23/1/95 <NA>
19: 3073 0.6 _leptosta. 91248°W 15241’N 1000 m 23/1/95 <NA>
20: 3074 3.9 vulgaris 91247'W 15240°N 1180 m 23/1/95 <NA>
21: 3075 4.0 vulgaris 91942"W 15939°N 1540 m 23/1/95 <NA>
22: 3076 6.4 coccineus 91231’W 14249’°N 2370 m 24/1/95 <NA>
23: 3077 9.5 coccineus 91229°W 14248°N 2070 m 24/1/95 <NA>
24: 3078 64 coccineus 91229°W 14247N 2050 m 24/1/95 <NA>
25: 3079 11.8 vulgaris 91229’W 14247'N 2050 m 24/1/95 <NA>
26: 3080 10.5 lunatus 91230°W 14246’N 1750 m 24/1/95 <NA>
27: 3081 6.9 vulgaris 91231’°W 14245’N 1730 m 24/1/95 <NA>
V1 V2 V3 V4 V5 V6 V7 V8 V9