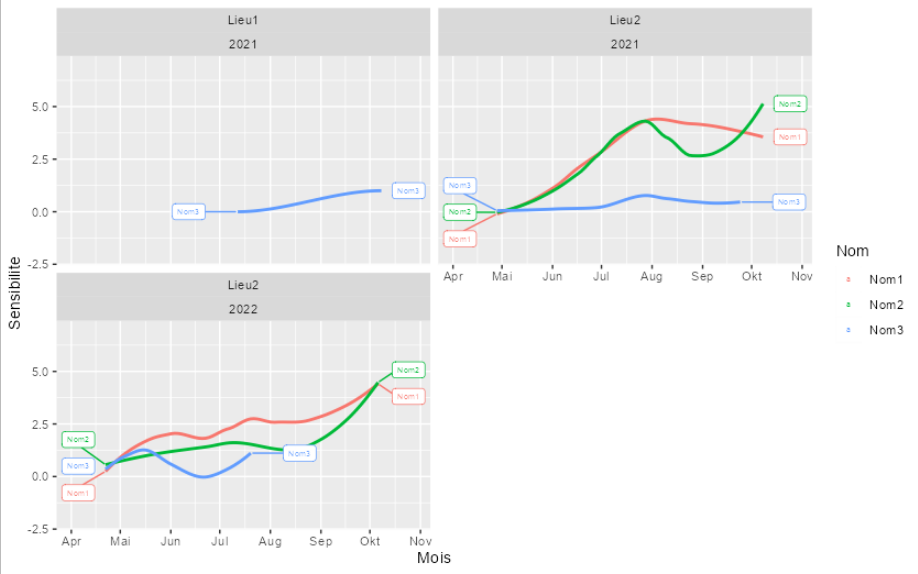
My goal is to label each curve drawn with geom_smooth at the end of the curve but when I use facet_wrap, the labels are not placed correctly and does not always appear.
This is my example :
library(ggrepel)
library(tidyverse)
df<-structure(list(month_day = structure(c(19124, 19124, 19132, 19132,19145, 19146, 19171, 19172, 19160, 19185, 19201, 19214, 19229,19244, 19110, 19259, 19273, 19104, 19116, 19130, 19131, 19144, 19166, 19179, 19193, 19208, 19229, 19243, 19256, 19271, 19131, 19145, 19145, 19171, 19171, 19157, 19160, 19185, 19185, 19201, 19201, 19214, 19214, 19229, 19229, 19244, 19244, 19110, 19110, 19259, 19259, 19273, 19273, 19104, 19104, 19116, 19144, 19130, 19166, 19179, 19208, 19229, 19243, 19256, 19256, 19271, 19271,19131, 19145, 19145, 19171, 19171, 19157, 19185, 19185, 19201, 19201, 19214, 19214, 19214, 19229, 19229, 19244, 19244, 19110, 19110, 19259, 19259, 19273, 19273, 19104, 19104, 19116, 19116,19130, 19144, 19166, 19179, 19179, 19193), class = "Date"),
Sensibilite = c(4,1, 0, 1, 0, 2, 3, 3, 2, 3, 4, 5, 5, 2, 0, 4, 4, 0, 1, 2, 3, 1,2, 2, 3, 3, 2, 2, 5, 4, 0, 2, 1, 3, 0, 2, 1, 5, 4, 5, 5, 5, 1, 4, 1, 3, 0, 0, 0, 5, 4, 5, 5, 0, 1, 1, 1, 1, 2, 1, 2, 2, 0, 4,3, 5, 4, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 2, 0, 0, 0, 0, 1, 0, 0,0, 1, 0, 1, 1, 1, 0, 1, 0, 2, 0, 0, 0, 1, 1),
Nom = c("Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom1","Nom1", "Nom1", "Nom1", "Nom1", "Nom1", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2","Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2","Nom2", "Nom2", "Nom2", "Nom2","Nom2", "Nom2", "Nom2", "Nom2","Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom2", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3", "Nom3","Nom3", "Nom3"),
Annee = c("2021","2021","2021", "2021", "2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2022", "2022", "2022","2022", "2022","2022", "2022", "2022", "2022", "2022","2022", "2022","2022","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021","2021", "2021","2021","2021","2021","2021", "2021","2021","2021","2021","2022","2022","2022","2022","2022","2022","2022","2022","2022","2022","2022","2022", "2022","2022","2021","2021","2021","2021","2021","2021", "2021","2021","2021","2021","2021","2021","2021","2021","2021","2021", "2021","2021","2021","2021","2021","2021", "2021","2022","2022","2022", "2022","2022", "2022","2022","2022","2022", "2022"),
Lieu = c("Lieu1","Lieu1","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu1","Lieu1","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu1","Lieu1","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2","Lieu2" )),
row.names = c(NA, -100L), class = c("tbl_df", "tbl", "data.frame"))
ggplot(df, aes(month_day, Sensibilite, label = Nom, color = Nom))
geom_smooth(se = FALSE)
geom_label_repel(aes(label = after_stat(ifelse(x %in% range(x), color, NA_character_))),
stat = "smooth",
max.overlaps = Inf,size = 2)
facet_wrap(Lieu~Annee,ncol = 2)
scale_y_continuous(limits = c(0,5))
scale_x_date(expand = expansion(mult = 0.25),name = "Mois",date_breaks = "1 month",date_labels = "%b")
when I remove
facet_wrap(Lieu~Annee,ncol = 2)
the labels are correct (even if I would prefer them only at the end of the curve)
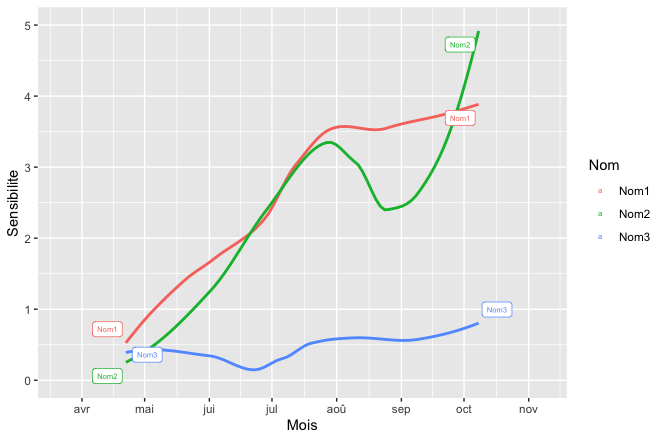
but as soon as I put this line again, some labels appears at the start of the curve, at the end, or even not at all.
Does somebody have an idea how I can fix this ?
CodePudding user response:
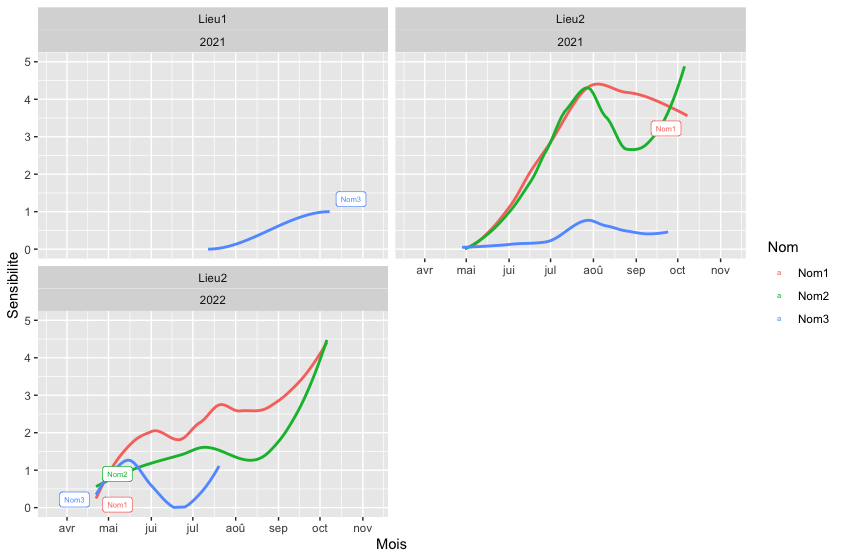
The first issue is that in your ifelse you did not take into account the grouping by color and PANEL (i.e. facetting). To this end I added a custom helper function which using a bit of dplyr sets the label per color and PANEL.
To fix your second issue I use two geom_label_repel layers to add the labels separately at the lower and the upper end. Doing so allows to nudge the labels at the lower end to the left and to the right at the upper end. Also I set the direction to shift or repel the labels to the "y" direction. Finally to make room for the labels I expand the x scale additively by the same amount used to nudge the labels.
library(ggrepel)
library(tidyverse)
helper_label <- function(x, color, PANEL, FUN = "min") {
FUN <- switch(FUN,
"min" = min,
"max" = max
)
data.frame(x, color, PANEL) %>%
group_by(PANEL, color) %>%
mutate(label = ifelse(x %in% FUN(x), color, NA_character_)) %>%
ungroup() %>%
pull(label)
}
ggplot(df, aes(month_day, Sensibilite, label = Nom, color = Nom))
geom_smooth(se = FALSE)
geom_label_repel(aes(label = after_stat(helper_label(x, color, PANEL, "min"))),
stat = "smooth",
max.overlaps = Inf, size = 2, na.rm = TRUE, direction = "y", nudge_x = -30
)
geom_label_repel(aes(label = after_stat(helper_label(x, color, PANEL, "max"))),
stat = "smooth",
max.overlaps = Inf, size = 2, na.rm = TRUE, direction = "y", nudge_x = 30
)
facet_wrap(Lieu ~ Annee, ncol = 2)
scale_x_date(
expand = expansion(add = 30), name = "Mois",
date_breaks = "1 month", date_labels = "%b"
)