Using: the package "glmnet"
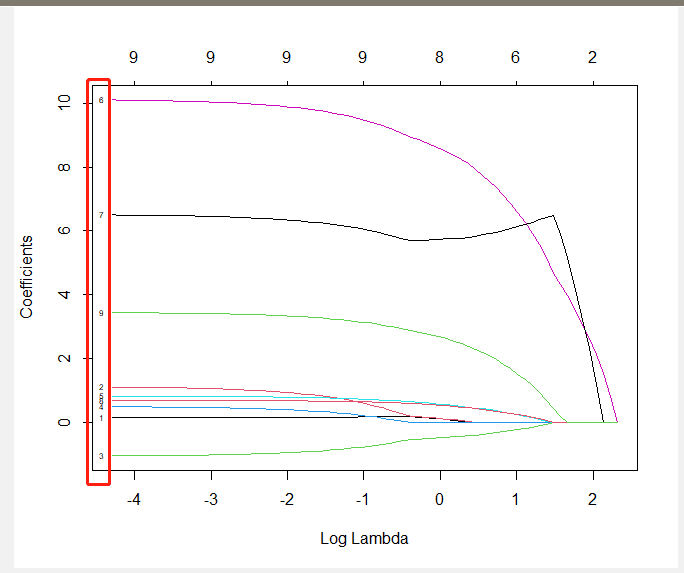
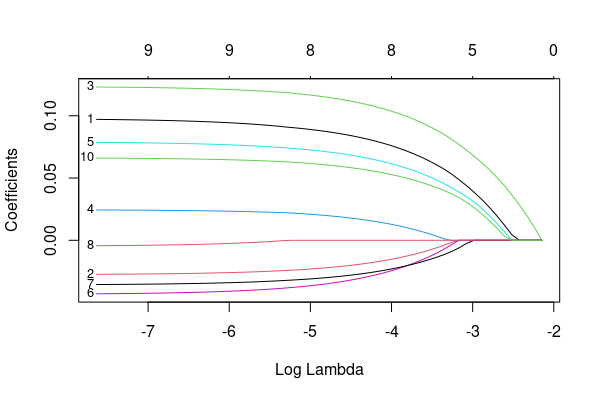
Problem: I use the plot function to plot a lasso image, I feel the labels are too small. So, I want to change the cex, but, it's not working. I looked up the documents of "glmnet", the plot function seems like normal plot. Any idea?
My code:
plot(f1, xvar='lambda', label=TRUE, cex=1.5)
Other: I tried like cex.lab=3. It worked but for the x-y axis labels.
CodePudding user response:
What you are actually using is the generic plot which dispatches a method depending on the class of the object. In this case,
class(fit1)
# [1] "elnet" "glmnet"
glmnet:::plot.glmnet will be used, which internally uses glmnet:::plotCoef. And here lies the problem; in glmnet:::plotCoef the respective cex parameter in glmnet:::plotCoef is hard coded to 0.5.―We need a hack:
plot.glmnet <- function(x, xvar=c("norm", "lambda", "dev"),
label=FALSE, ...) {
xvar <- match.arg(xvar)
plotCoef2(x$beta, lambda=x$lambda, df=x$df, dev=x$dev.ratio, ## changed
label=label, xvar=xvar, ...)
}
plotCoef2 <- function(beta, norm, lambda, df, dev, label=FALSE,
xvar=c("norm", "lambda", "dev"), xlab=iname, ylab="Coefficients",
lab.cex=0.5, xadj=0, ...) {
which <- glmnet:::nonzeroCoef(beta)
nwhich <- length(which)
switch(nwhich 1, `0`={
warning("No plot produced since all coefficients zero")
return()
}, `1`=warning("1 or less nonzero coefficients; glmnet plot is not meaningful"))
beta <- as.matrix(beta[which, , drop=FALSE])
xvar <- match.arg(xvar)
switch(xvar, norm={
index=if (missing(norm)) apply(abs(beta), 2, sum) else norm
iname="L1 Norm"
approx.f=1
}, lambda={
index=log(lambda)
iname="Log Lambda"
approx.f=0
}, dev={
index=dev
iname="Fraction Deviance Explained"
approx.f=1
})
dotlist <- list(...)
type <- dotlist$type
if (is.null(type))
matplot(index, t(beta), lty=1, xlab=xlab, ylab=ylab, type="l", ...)
else matplot(index, t(beta), lty=1, xlab=xlab, ylab=ylab, ...)
atdf <- pretty(index)
prettydf <- approx(x=index, y=df, xout=atdf, rule=2, method="constant", f=approx.f)$y
axis(3, at=atdf, labels=prettydf, tcl=NA)
if (label) {
nnz <- length(which)
xpos <- max(index)
pos <- 4
if (xvar == "lambda") {
xpos <- min(index)
pos <- 2
}
xpos <- rep(xpos xadj, nnz) ## changed
ypos <- beta[, ncol(beta)]
text(xpos, ypos, paste(which), cex=lab.cex, pos=pos) ## changed
}
}
If we load the two hacked functions, we now can adapt the coefficient labels. New are the parameters lab.cex for size and xadj to adjust the x position of the numbers.
plot(fit1, xvar='lambda', label=TRUE, lab.cex=.8, xadj=.085)
Data:
set.seed(122873)
x <- matrix(rnorm(100 * 10), 100, 10)
y <- rnorm(100)
fit1 <- glmnet(x, y)