I am using Images.save() to save a file locally with img. So In one repo, Images.save works but in another one, it doesn't.
Here is the work 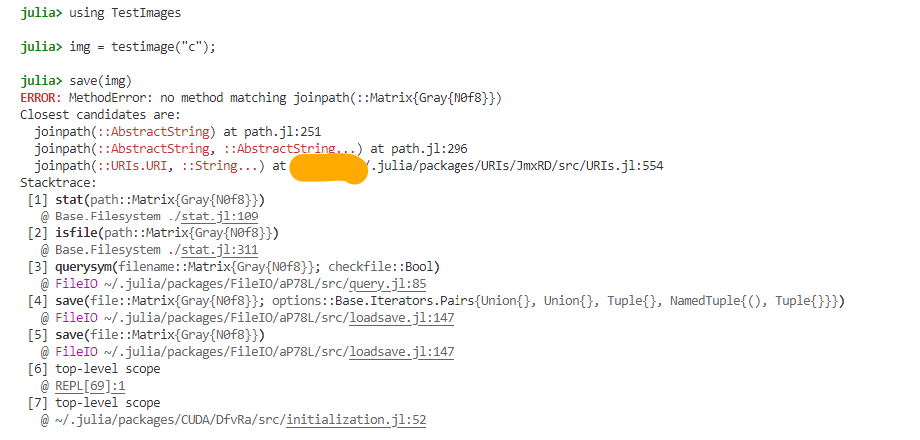
using TestImages
img = testimage("c");
save(img)
I ran a @which save, the output is FileIO. Not sure why it picks joinpath!
UPDATE: So the issue is with GaussianMixtureModels library which over writes FileIO.save. Not sure how to work around it?
CodePudding user response:
Issue for a start is incorrect argument use for the saving method. You can find more details about arguments of a specific function using ?save in REPL, it will show all possible methods and the arguments needed.
julia> using TestImages, FileIO
[ Info: Precompiling TestImages [5e47fb64-e119-507b-a336-dd2b206d9990]
julia> img = testimage("c");
[ Info: Precompiling TiffImages [731e570b-9d59-4bfa-96dc-6df516fadf69]
julia> save("test.png", img)
[ Info: Precompiling PNGFiles [f57f5aa1-a3ce-4bc8-8ab9-96f992907883]
julia> size(img)
(512, 512)
help?> save
search: save savestreaming add_saver StackOverflowError AbstractVector
• save(filename, data...) saves the contents of a
formatted file, trying to infer the format from
filename.
• save(Stream{format"PNG"}(io), data...) specifies the
format directly, and bypasses the format query.
• save(File{format"PNG"}(filename), data...) specifies the
format directly, and bypasses the format query.
• save(f, data...; options...) passes keyword arguments on
to the saver.
Like Dan mentioned in comments, I am able to save too without issues. Can you please share details provided by versioninfo() or perhaps try to update to latest version of Julia and the Testimages.jl.
CodePudding user response:
UPDATE: So the issue is with GaussianMixtureModels library which over writes FileIO.save. Not sure how to work around it?
In Julia programming, There might be at least one common function among the imported modules. For example, both DataFrames and Pandas share a function named DataFrame. When a user tries to make a DataFrame there might be a problem (ambiguity and confusion) with which DataFrame to use for this purpose (another important thing here is that in such situations the functions might have different arguments in case of type or count). Hence, in these cases, it's better to call the function as a suffix of the desired module.
In your case, you're dealing with the save function, and apparently, there is more than one module that shares the function. Then prefixing the name of the module hopefully would solve the problem. For example, if you want to save the img, then it's probably efficient to write FileIo.save(img).
But, be cautious and first try to observe the methods of the function by methods(FileIo.save). This will help you to provide the arguments that are needed for using the function in the related module.
Another workaround would be utilizing the import keyword rather than using. This kind of importing forces the user to prefix the name of the module to call a function from:
julia> import Pandas as pd
julia> using DataFrames
julia> methods(DataFrame)
This will result in 26 methods (considering the latest version of DataFrames so far) that all point to a location: ...\.julia\packages\DataFrames. This means in this case each time a user writes DataFrame the compiler chooses from the locally available functions. But, if someone needs to use the DataFrame function of the Pandas module, then they should write pd.DataFrame (which isn't actually common among the Julia programmers.)
Another options is to write this (which is more preferable):
julia> using DataFrames
julia> using Pandas: DataFrame as dtf
Then there will be no ambiguity when you call the DataFrame function and is homogeneous with the Julia programming style.
Finally, I highly recommend you to read the official doc until the end around this matter.

