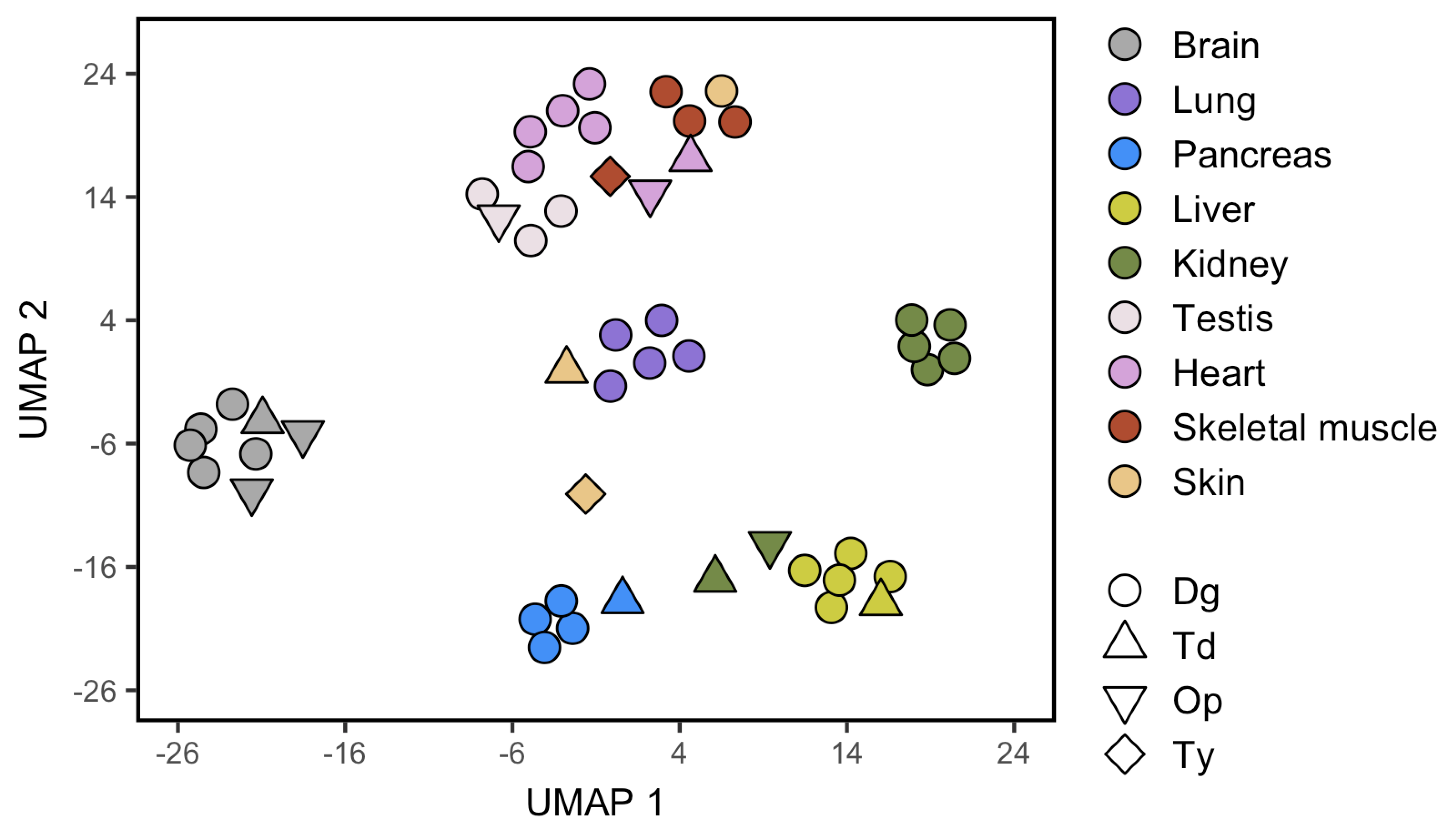
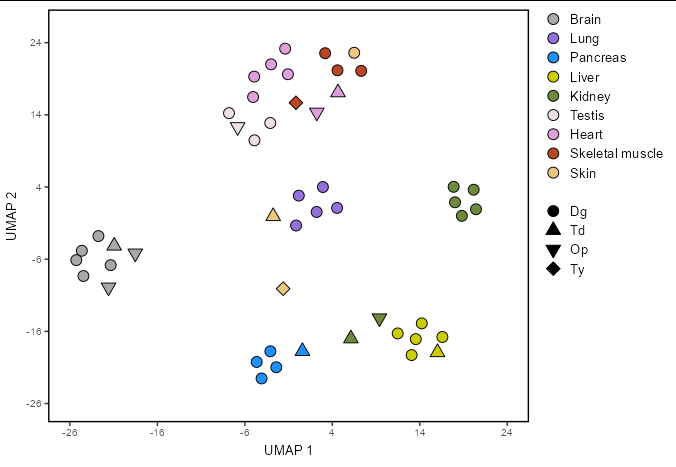
I am struggling to get the final bit of thing I need to complete one plot.
This is the tale:
UMAP1 UMAP2 color tissue shape
BR1.RPM. 24.4555644927102 8.33081550406305 darkgrey Brain 21
BR2.RPM. 24.6344541391525 4.82860000294483 darkgrey Brain 21
BR3.RPM. 21.3411330596164 6.82626266175396 darkgrey Brain 21
BR4.RPM. 25.2706589831506 6.12795744057223 darkgrey Brain 21
BR5.RPM. 22.7412144591854 2.79845872023771 darkgrey Brain 21
HE1.RPM. 4.93012565944078 -19.2942174526575 plum Heart 21
HE2.RPM. 2.99869262617741 -20.9994250048535 plum Heart 21
HE3.RPM. 1.07403447412913 -19.6200772829839 plum Heart 21
HE4.RPM. 5.05658841417385 -16.4676995230271 plum Heart 21
HE5.RPM. 1.38979293698651 -23.1667225134936 plum Heart 21
KI1.RPM. -18.8116579138644 -0.00462977244554885 darkolivegreen4Kidney 21
KI2.RPM. -20.432681992004 -0.915727298879803 darkolivegreen4Kidney 21
KI3.RPM. -18.030857579405 -1.88453627174062 darkolivegreen4Kidney 21
KI4.RPM. -17.8739263530499 -4.03002130738062 darkolivegreen4Kidney 21
KI5.RPM. -20.1634636202346 -3.6263638737022 darkolivegreen4Kidney 21
LI1.RPM. -11.4770920275156 16.2842167192584 yellow3 Liver 21
LI2.RPM. -14.2305178863485 14.8920278578934 yellow3 Liver 21
LI3.RPM. -13.0721198535928 19.2812569438666 yellow3 Liver 21
LI4.RPM. -16.5863672861391 16.7787928871763 yellow3 Liver 21
LI5.RPM. -13.5477499873499 17.07104133669 yellow3 Liver 21
LU1.RPM. -2.92494582302321 -3.99929775102582 mediumpurple Lung 21
LU2.RPM. -4.55004758488599 -1.10444902760826 mediumpurple Lung 21
LU3.RPM. -2.21046905247079 -0.543716793747593 mediumpurple Lung 21
LU4.RPM. -0.171714494316746 -2.80912609668774 mediumpurple Lung 21
LU5.RPM. 0.138361668949329 1.33912348982787 mediumpurple Lung 21
PA1.RPM. 2.40256627188632 20.972474918705 dodgerblue Pancrea21
PA2.RPM. 3.07832860321791 18.7632129867164 dodgerblue Pancreas 21
PA3.RPM. 4.64721910111144 20.240083817214 dodgerblue Pancrea21
PA5.RPM. 4.08614060978053 22.523507524782 dodgerblue Pancrea21
SKI.RPM. -6.50616313409082 -22.5987512552561 #F0C57F Skin 21
SM1.RPM. -3.18280536469006 -22.5368142358323 #BC4524 Skeletal muscle 21
SM2.RPM. -4.5943110200305 -20.1688133359027 #BC4524 Skeletal muscle 21
SM3.RPM. -7.30641518202577 -20.0859051130635 #BC4524 Skeletal muscle 21
TE1.RPM. 7.80910486885204 -14.23430686181 lavenderblush2 Testis 21
TE2.RPM. 4.90001913797227 -10.4702885523713 lavenderblush2 Testis 21
TE3.RPM. 3.09328325061247 -12.8659248177456 lavenderblush2 Testis 21
Brain_Sha 20.9386190008756 4.15082143059336 darkgrey Brain 24
Heart_Sha -4.64687807513027 -17.0889913553132 plum Heart 24
Kidney_Sha -6.12575586753639 17.0038176223978 darkolivegreen4Kidney 24
Liver_Sha -16.0289300026197 18.9145544776388 yellow3 Liver 24
Pancreas_Sha -0.584268732931413 18.7479385055046 dodgerblue Pancreas 24
Skin_Sha 2.76066649656992 0.0628225671099258 #F0C57F Skin 24
Brain_Mdo 21.5843973523696 9.8934422431328 darkgrey Brain 25
Cerebellum_Mdo 18.5294231342495 5.18064808162601 darkgrey Brain 25
Heart_Mdo -2.23047530413099 -14.3377907628058 plum Heart 25
Kidney_Mdo -9.39418037560359 14.1691863077575 darkolivegreen4Kidney 25
Testis_Mdo 6.82340577182029 -12.3274677871282 lavenderblush2 Testis 25
Tcy_muscle 0.159978911767435 -15.6754517794312 #BC4524 Skeletal muscle 23
Tcy_skin 1.61794178327545 10.0875748984971 #F0C57F Skin 23
And this is the code I use:
UMAP$color = factor(UMAP$color, levels=c("darkgrey", "mediumpurple", "dodgerblue", "yellow3", "darkolivegreen4",
"lavenderblush2", "plum", "#BC4524", "#F0C57F"))
UMAP$tissue = factor(UMAP$tissue, levels=c("Brain", "Lung", "Pancreas", "Liver", "Kidney", "Testis", "Heart",
"Skeletal muscle", "Skin"))
UMAP$shape = factor(UMAP$shape, levels=c(21,24,25,23))
UMAP_plot = ggplot(UMAP, aes(x=-UMAP1, y=-UMAP2, fill=color, shape=shape))
geom_point(size=3.5)
scale_fill_manual(labels = c("Brain", "Lung", "Pancreas", "Liver","Kidney", "Testis", "Heart",
"Skeletal muscle", "Skin"),
values = c("darkgrey", "mediumpurple", "dodgerblue", "yellow3", "darkolivegreen4",
"lavenderblush2", "plum", "#BC4524", "#F0C57F"))
scale_shape_manual(labels = c("Dg","Td", "Op", "Ty"),
values = c(21, 24, 25, 23))
guides(fill = guide_legend(ncol=2))
ylim(c(-26,26)) xlim(c(-26,26))
xlab("UMAP 1") ylab("UMAP 2")
scale_y_continuous(limits = c(-26, 26), breaks = seq(-26, 26, 10))
scale_x_continuous(limits = c(-26, 24), breaks = seq(-26, 24, 10))
theme_bw()
theme(legend.position = "right", legend.title = element_blank(), legend.text = element_text(size=10),
legend.justification ="top")
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
theme(axis.text.x = element_text(size=8),
axis.text.y = element_text(size=8),
axis.title.x =element_text(size=10, margin = margin(t = 5)),
axis.title.y =element_text(size=10, margin = margin(r = 5)),
plot.margin = margin(t=0.25, b=0.25, l=0.25, r=0.25, "cm"))
theme(legend.key.size = unit(1, 'lines'))
theme(panel.border = element_rect(linetype = "solid", size=1, color="black"))
guides(fill = guide_legend(override.aes =list(shape = 21), nrow=10))
theme(legend.spacing.y = unit(0,"lines"),
legend.margin = margin(0,0,0,0))
Which generates the following plot:
I would like to put the shape legend in black color, not as it is now with void shapes. I have tried several ways to add the color to that legend specifically but I always end up putting all the dots in black...
Is there a way to manually set black color for only the scale_shape_manual option?
Is there an alternative code to generate the desired plot maybe??
Many thanks
CodePudding user response:
You can add fill = "black" to the override.aes of the shape guide. As a little tip, keeping your code organised into logical sections while minimizing duplicated calls really helps to focus debugging.
ggplot(UMAP, aes(-UMAP1, -UMAP2, fill = color, shape = shape))
geom_point(size = 3.5)
scale_fill_manual(labels = levels(UMAP$tissue),
values = levels(UMAP$color),
guide = guide_legend(override.aes = list(shape = 21),
nrow = 10))
scale_shape_manual(labels = c("Dg","Td", "Op", "Ty"),
values = c(21, 24, 25, 23),
guide = guide_legend(override.aes =list(fill = "black")))
scale_y_continuous("UMAP 2", limits = c(-26, 26), breaks = seq(-26, 26, 10))
scale_x_continuous("UMAP 1", limits = c(-26, 24), breaks = seq(-26, 24, 10))
theme_bw()
theme(legend.title = element_blank(),
legend.text = element_text(size = 10),
legend.justification = "top",
legend.spacing.y = unit(0, "lines"),
legend.margin = margin(0, 0, 0, 0),
legend.key.size = unit(1, 'lines'),
panel.border = element_rect(size = 1, color = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.text = element_text(size = 8),
axis.title.x = element_text(size = 10, margin = margin(t = 5)),
axis.title.y = element_text(size = 10, margin = margin(r = 5)),
plot.margin = margin(t = 0.25, b = 0.25, l = 0.25, r = 0.25, "cm"))