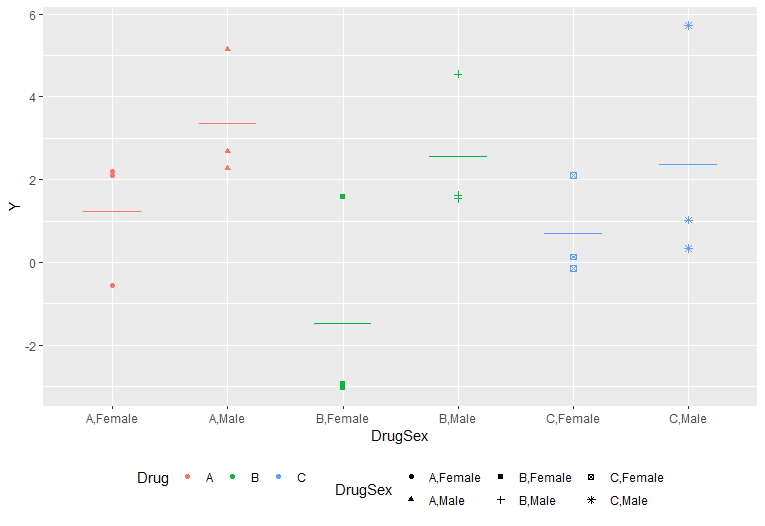
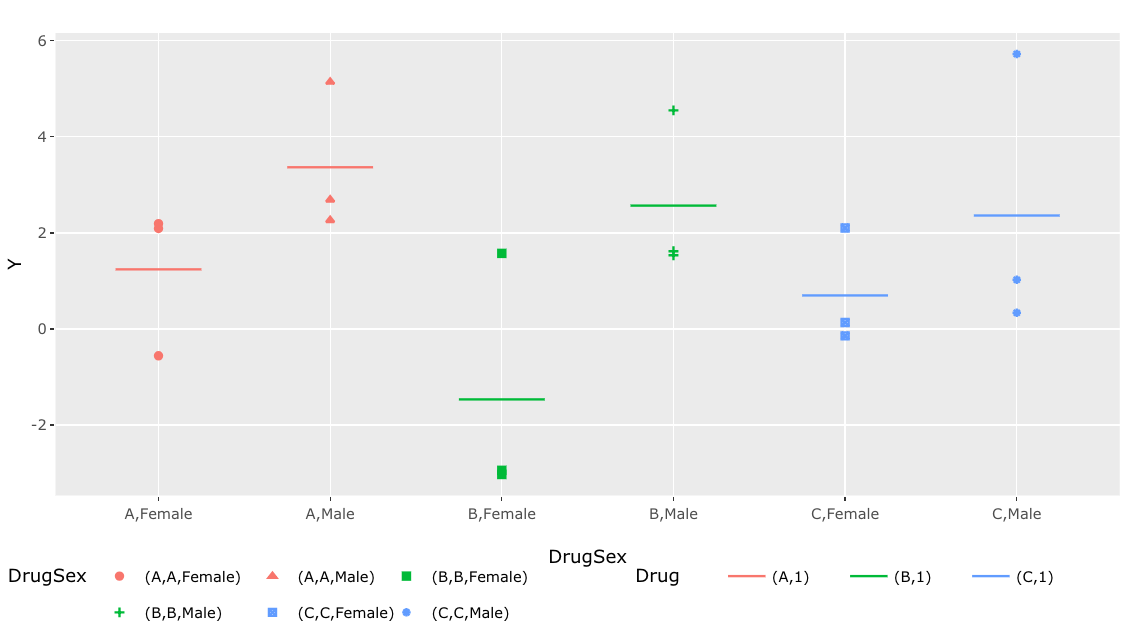
Using the ggplotly function from the plotly package, I am making the ggplot output interactive. When I use the ggplotly function to a ggplot-generated plot, however, the legend style and data shapes are completely altered. How can I use plotly while maintaining the shapes and legends as the primary plot?
data:
"","Day","Drug","Sex","Y","DrugSex"
"1",1,"A","Female",2.192306074,"A,Female"
"2",1,"B","Male",4.551912798,"B,Male"
"3",1,"B","Female",1.574070652,"B,Female"
"4",1,"C","Female",-0.143946163,"C,Female"
"5",1,"A","Male",5.144422967,"A,Male"
"6",1,"C","Male",5.724705829,"C,Male"
"7",2,"A","Male",2.691617258,"A,Male"
"8",2,"B","Female",-3.0289955,"B,Female"
"9",2,"C","Male",0.338102762,"C,Male"
"10",2,"A","Female",-0.558581233,"A,Female"
"11",2,"B","Female",-2.942620032,"B,Female"
"12",2,"C","Male",1.024670497,"C,Male"
"13",3,"A","Male",2.264980803,"A,Male"
"14",3,"C","Female",2.103722883,"C,Female"
"15",3,"A","Female",2.091621938,"A,Female"
"16",3,"B","Male",1.535299922,"B,Male"
"17",3,"B","Male",1.618399767,"B,Male"
"18",3,"C","Female",0.136160703,"C,Female"
After copying you may need to run the following command to convert it to the dataframe:
df <- read.delim("clipboard", sep = ",")
Here is the data using the dput function:
df <-structure(list(Day = c(1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L), Drug = c("A", "B", "B", "C",
"A", "C", "A", "B", "C", "A", "B", "C", "A", "C", "A", "B", "B",
"C"), Sex = c("Female", "Male", "Female", "Female", "Male", "Male",
"Male", "Female", "Male", "Female", "Female", "Male", "Male",
"Female", "Female", "Male", "Male", "Female"), Y = c(2.192306074,
4.551912798, 1.574070652, -0.143946163, 5.144422967, 5.724705829,
2.691617258, -3.0289955, 0.338102762, -0.558581233, -2.942620032,
1.024670497, 2.264980803, 2.103722883, 2.091621938, 1.535299922,
1.618399767, 0.136160703), DrugSex = structure(c(1L, 4L, 3L,
5L, 2L, 6L, 2L, 3L, 6L, 1L, 3L, 6L, 2L, 5L, 1L, 4L, 4L, 5L), levels = c("A,Female",
"A,Male", "B,Female", "B,Male", "C,Female", "C,Male"), class = "factor")), row.names = c(NA,
-18L), class = "data.frame")
Here is the code for generating the plot:
df_means <- df %>% group_by(DrugSex) %>%
summarise(color = first(Drug),Mean = mean(Y)) %>%
rename(grouping_var = 1) %>% mutate(x = seq(length(unique(grouping_var))))
p <-df %>% ggplot(aes(x = DrugSex, y = Y))
geom_point(aes(color = Drug, shape = DrugSex))
geom_segment(data = df_means, aes(x=x-0.25, xend=x 0.25, y=Mean, yend=Mean, color=
color),inherit.aes = F, show.legend = F)
theme(legend.position = 'bottom',
legend.key=element_blank() #transparent legend panel
)
I want my plotly to look like the p
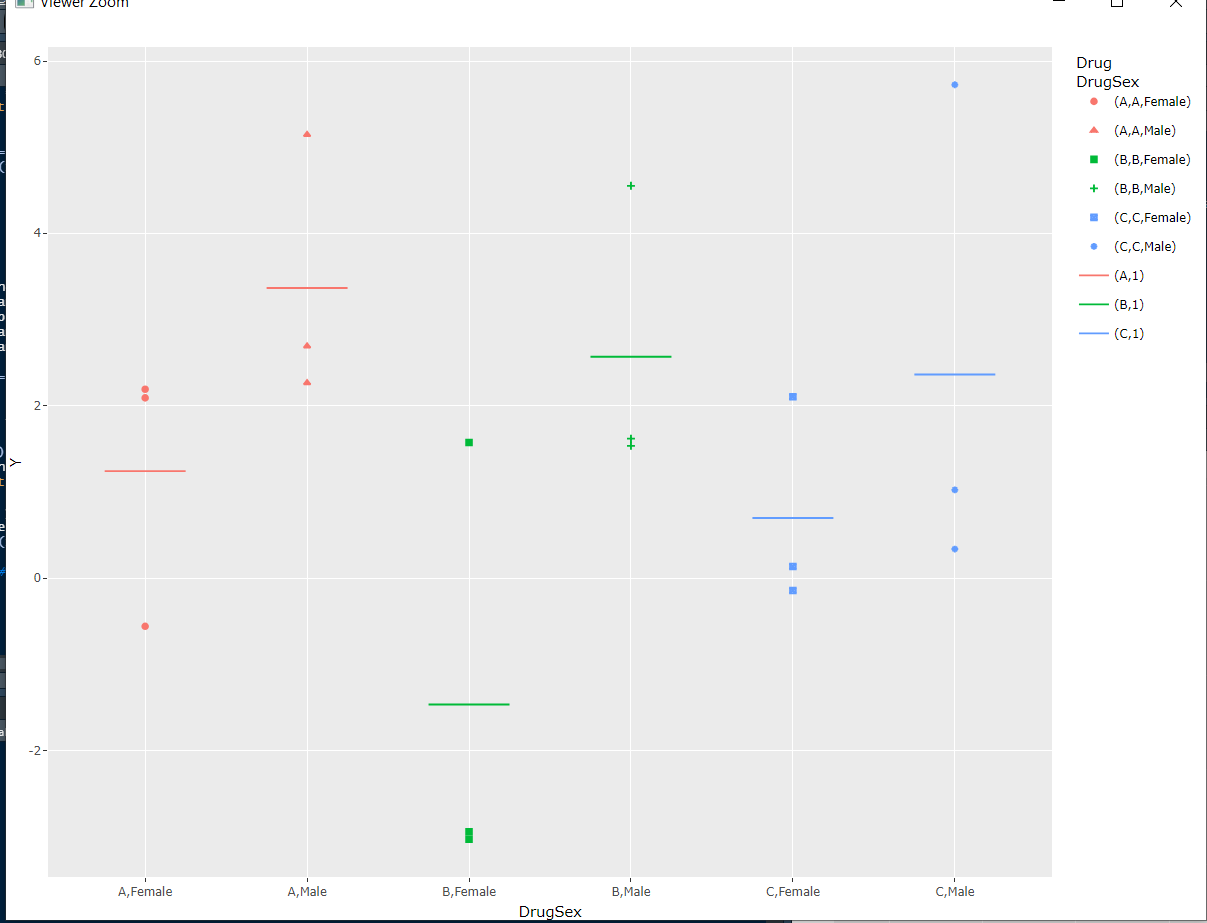
However, after executing ggplotly(p), it appears as follows:
CodePudding user response:
I've got two methods you can work with. Both methods will eliminate the ability to use pointer events in the legend (where you click on the legend to show or hide information).
One method strictly uses R, has many steps, and is somewhat complicated.
The other method uses htmlwidgets::onRender and Javascript. This method is almost no work on your part.
The method that is strictly R does require finessing. It's also based on the assumption that you're using RStudio. It might work in other IDEs, but I'm not sure. Essentially you'll capture what's shown in the Viewer pane at one point. I have no idea how big your viewer pane is, so I can't tell you the numbers to use.
I've added a function that changes some Plotly defaults. For example, when you render into your browser or the viewer pane, it is set to fill = True (which will likely run your legend off the viewable area of the webpage). I've set the height to 80% to ensure the legend is shown. When we create the plot, this function is piped at the end. (You'll see it used in the code.)
fixHt <- function(plt) {
plt <- plotly_build(plt) # ensure data object is in plt object
plt$sizingPolicy$defaultHeight <- '100%'
plt$sizingPolicy$padding <- '15'
plt$sizingPolicy$browser$defaultWidth <- '100vw' # 100% viewer width
plt$sizingPolicy$browser$defaultHeight <- '100vh' # 100% viewer height
plt$sizingPolicy$browser$fill <- F
plt$sizingPolicy$viewer$fill <- F
plt # return the plotly object with these changes
}
Method 1: Strictly R
We'll create a second plot. Don't change your current plot; create a new one. In the new plot, we'll exaggerate the legend. The idea is to get a better resolution and to make the text relatively sized (versus the typical rendering).
In the end, a picture of your ggplot2 legend will be added to your plot.
library(plotly)
library(cowplot)
library(magick)
# if using within R tools only --
# start with enhancing the legend for clarity in transfer
p1 <- df %>% ggplot(aes(x = DrugSex, y = Y))
geom_point(aes(color = Drug, shape = DrugSex))
geom_segment(data = df_means, aes(x=x-0.25, xend=x 0.25, y=Mean, yend=Mean, color=
color),inherit.aes = F, show.legend = F)
theme(legend.position = 'bottom',
legend.key=element_blank(), #transparent legend panel
legend.key.size = unit(.5, 'cm'),
legend.title = element_text(size = rel(1.2)), # relative text size
legend.text = element_text(size = rel(1))) # relative text size
Using cowplot, capture the legend. If you want to see it, you can use cowplot::plot_grid(pLeg) to print it to the plot pane.
pLeg <- get_legend(p1) # capture the legend
The next step will require you to watch exactly what happens in the Viewer pane. The code between dev.off and dev.off may have re-run a few times. (It depends on your settings.) I've added quite a few comments here that would behoove you to read and heed.
Essentially, you'll print the legend to the viewer pane while magick's image_graph is listening. Whatever you see in the viewer pane will end up in the object img.
If when you run dev.off you see
Error in dev.off() : cannot shut down device 1 (the null device)
That means that you've cleared out everything. It means the same thing as when you see
null device
1
When you run this code, I suggest that you make your viewer pane really wide. You can move it back after dev.off.
# clear plot & viewer pane if not already**
dev.off() # should say 'null device'
# if not re-exacute until it does
# if it says it 'can't`, continue
autoviewer_enable()
# super high res, reduce in plot; so plot resized still has clear legend
img <- magick::image_graph(res = 125, height = 45)
# entire legend must show in the viewer pane;
# what you see is what you get
# chances are you need to make the viewer pane bigger between the two dev.off calls
cowplot::plot_grid(pLeg2)
autoviewer_disable()
dev.off() # clear out Magick control of Viewer
img is now an image that can be manipulated with the magick library. Next, trim the white space out from around the legend and then use as.raster and Plotly's raster2uri to prepare the image for Plotly.
Now that it's an image, you can simply run the object name to print it.
# remove background
img_t <- image_trim(img)
img_r <- raster2uri(as.raster(img_t)) # prepare for Plotly
It's ready for plotting. A few changes: hide the legend, add a margin to make space for the legend image, and add the legend image. Depending on your perspective and viewer pane size, you may want to adjust the numbers for x, y, sizex, and sizey in the call for images.
Since this is not in the plot, it's set to paper space. The left-to-right or bottom-to-top range is 0 to 1. That means that 0 is the leftmost or bottommost point of the viewing area, whereas .5 is the middle. In Plotly this is called the domain. (The default is 0 to 1, but you could actually set it to anything you want.)
Anchors in plotly are for alignment. If you want to place something at the point (0, 0), if you xanchor left, then the leftmost end is at the point (0, 0). if you xanchor center, then the middle is at (0, 0). yanchor works the same way.
sizex and sizey are multipliers. If the image is 100px by 100px and Plotly says it's 400 by 400 (even if your viewer pane is 1 by 1!!), then sizex set to 1, makes that object 1/4th the size of the viewing area. If sizex is set to 4, then the entire `viewer is that object. The easiest way to understand it is to change it a few times and see how it changes the plot. While changing it (for understanding), don't change the size of the viewer pane.
ggplotly(p) %>%
layout(
showlegend = F,
margin = list(t = 10, r = 10, b = 110, l = 20, pad = 4),
images = list(
list(
source = img_r, xref = 'paper', x = .5, xanchor = 'center',
yref = 'paper', y = -.3, sizex = .6, sizey = .6))) %>% fixHt()
In my current Viewer pane, this looks like this. The legend seems to small, right?
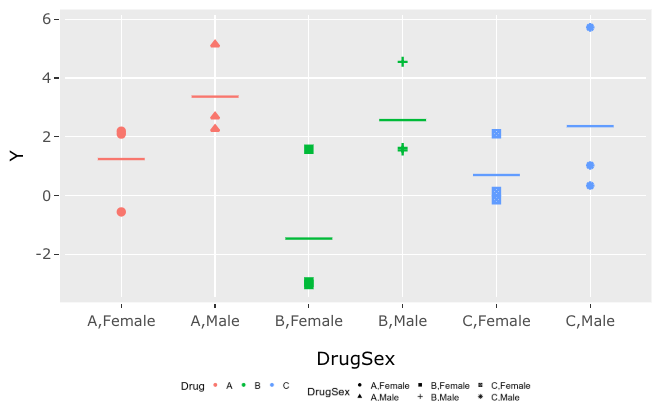
If I change my viewer pane, making it wider, this is what I get. The legend looks proportional.
Method 2: Using htmlwidgets::onRender
I didn't call the htmlwidgets library, I just appended it to the function. For this method, I changed none of your code. Nothing that was used in method 1 is used here.
Because of the transfer from ggplot to plotly, the text size is excessive. You may need to view this in the browser or expand your viewing pane. Alternatively, you can modify the text sizes in ggplot or in Plotly.
For example, if you wanted the ticktext size to match the legend item size an the xaxis title size to match the legend title size, you could use the following after naming the ggplotly object plt.
plt$x$layout$legend$font$size <- plt$x$layout$xaxis$tickfont$size
plt$x$layout$legend$title$font$size <- plt$x$layout$xaxis$title$font$size
You may want or need to adjust the y position of the legend in the R coding for layout. Even without the Javascript, you may have to adjust a legend positioned at the bottom, so it doesn't overlap the x-axis title.
I tried to add a lot of comments in the Javascript so that you could follow along if you wanted to understand what it did.
library(plotly)
ggplotly(p) %>%
layout(legend = list(orientation = "h", xanchor = "center",
x = .5, y = -.08)) %>% fixHt() %>%
htmlwidgets::onRender("
function(el, x) { /* starting with subenclosure in legend*/
gr = el.querySelector('g.legend rect.bg');
gr_w = gr.getAttribute('width');
gr_w2 = Number(gr_w)/2;
gr1 = gr.cloneNode(true);
gr1.setAttribute('width', gr_w2);
gr2 = gr1.cloneNode(true);
gr2.setAttribute('transform', 'translate(' gr_w2 ', 0)');
gr.parentElement.insertBefore(gr1, gr.parentElement.children[1]);
gr.parentElement.appendChild(gr2);
/* separate legend titles */
leg = el.querySelector('g.legend g.scrollbox'); /* capture legend */
l2 = leg.firstChild.cloneNode(true); /* deep copy legend titles */
leg.firstChild.innerHTML = leg.firstChild.firstChild.textContent; /* remove 2nd legend title */
l2.innerHTML = l2.children[1].textContent; /* 2nd leg title to 1st pos */
l2x = l2.getAttribute('x');
elw = .05 * el.clientWidth;
l2.setAttribute('x', Number(l2x) elw);
/* move first legend items to second row */
capTr = []; /* capture transforms of 2nd; 3rd leg items; global var*/
function mover(fc, tc, leg) { /* from child index; to child index; leg element */
if(fc == 1) {
leg.children[tc].setAttribute('transform', 'translate(0, 29)')
} else {
ft = leg.children[fc].getAttribute('transform'); /* capture xy translation */
/*capTr.push(ft);*/
tx = Number(ft.split(',')[0].replace(/^\\D /, '')); /* extract x transl */
capTr.push(tx);
leg.children[tc].setAttribute('transform', 'translate(' tx ', 29)');
}
}
/* moving elements within */
/* first is at 0, 0; (it's child 1 (legend name is child 0)) */
mover(1, 4, leg);
/* collect the 2:3 transforms to change the 5:6 legend items */
mover(2, 5, leg);
mover(3, 6, leg);
/* moving elements out */
els = ''; /* string to store the remaining legend nodes */
/* get minimum x val for transform after changing str to int */
posr = Math.min(...(capTr.map(function(str) {return parseInt(str);})));
for(i = 3; i > 0; i--) { /* move the Drug legend content*/
l2_el = leg.children[leg.children.length - i].cloneNode(true);
leg.removeChild(leg.children[leg.children.length - i]);
if(i == 3) {
l2_el.setAttribute('transform', 'translate(' elw ', 0)');
els = l2_el.outerHTML;
} else {
var val = [3, 2, 1][i];
l2_el.setAttribute('transform', 'translate(' (((posr * .8) * val) elw) ', 0)');
els = l2_el.outerHTML;
}
}
cpath = leg.getAttribute('clip-path'); /* consolidate the second legend */
ih = '<g class=\"scrollbox\" clip-path=\"' cpath '\"';
ih = ' transform=\"translate(' gr_w2 ', 0)\" >';
ih = l2.outerHTML els '</g>'; /* end g.scrollbox */
gr.parentElement.innerHTML = gr.parentElement.innerHTML ih;
$(window).on('resize', function(e) {
window.location.reload();
}); /* end event */
}")
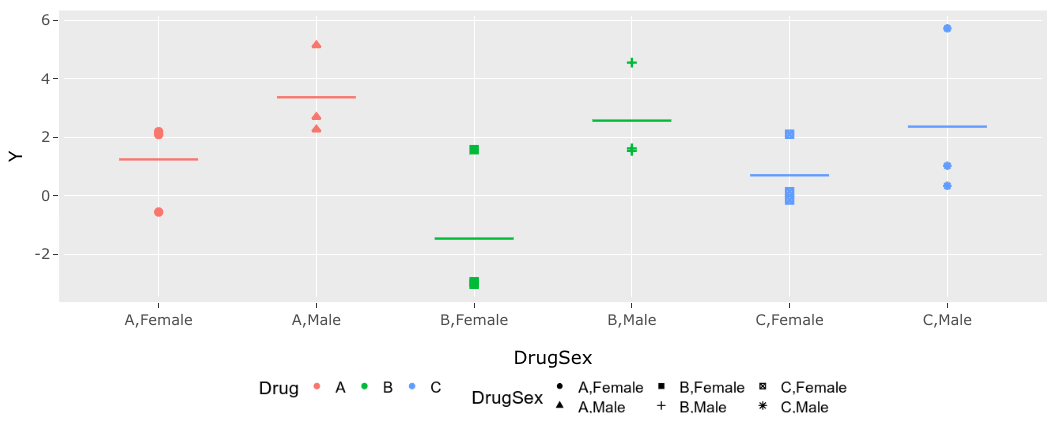
This image reflects the appearance if the font sizes are between the legend and the axes are matched.