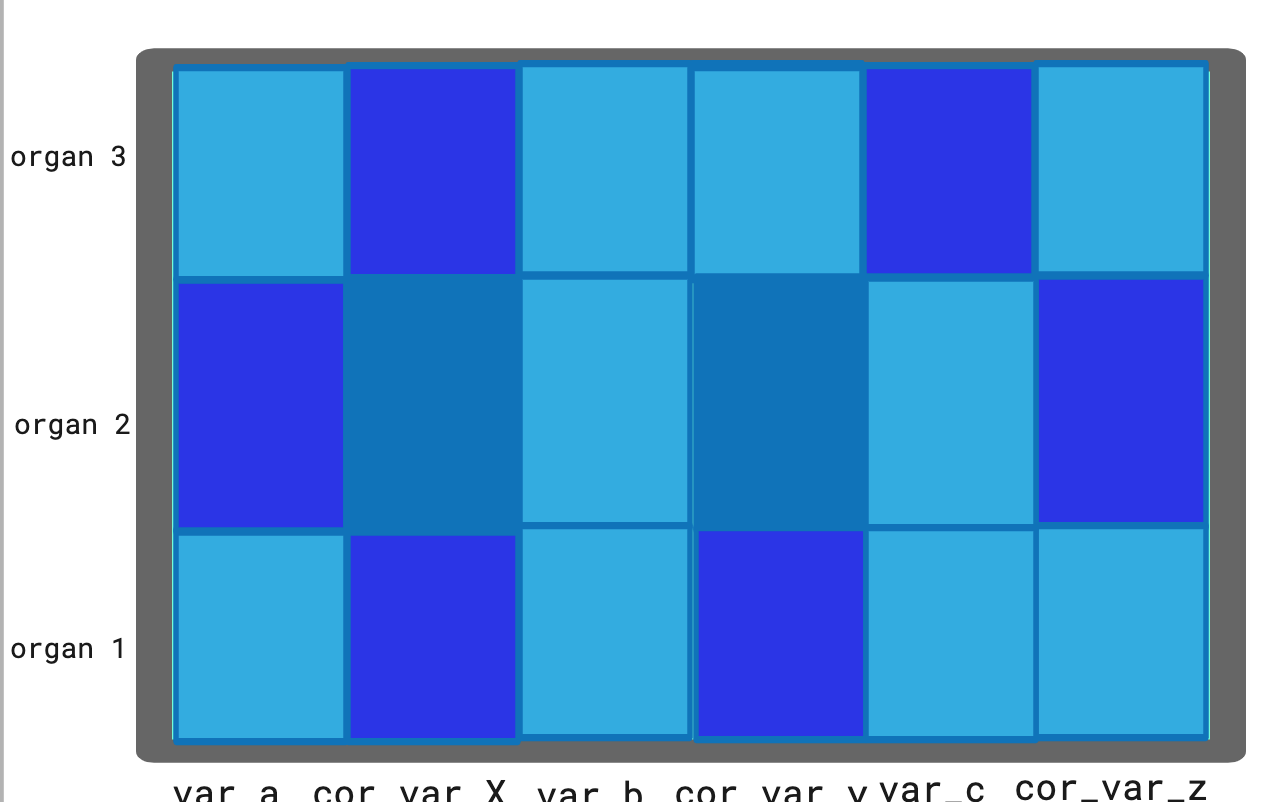
I'm trying to combine two heatmaps. I want var_a and var_x on the y axis with for example: var_a first and then var_x. I don't know if I should do this by changing the dataframe or combining them, or if I can do this in ggplot.
Below I have some example code and a drawing of what I want (since I don't know if I explained it right).
I hope someone has ideas how I can do this either in the dataframe or in ggplot!
Example code:
df_one <- data.frame(
vars = c("var_a", "var_b", "var_c"),
corresponding_vars = c("var_x", "var_y", "var_z"),
expression_organ_1_vars = c(5, 10, 20),
expression_organ_2_vars = c(50, 2, 10),
expression_organ_3_vars = c(5, 10, 3)
)
df_one_long <- pivot_longer(df_one,
cols=3:5,
names_to = "tissueType",
values_to = "Expression")
expression.df_one <- ggplot(df_one_long,
mapping = aes(y=tissueType, x=vars, fill = Expression))
geom_tile()
expression.df_one
df_two <- data.frame(
corresponding_vars = c("var_x", "var_y", "var_z"),
expression_organ_1_corresponding_vars = c(100, 320, 120),
expression_organ_2_corresponding_vars = c(23, 30, 150),
expression_organ_3_corresponding_vars = c(89, 7, 200)
)
df_two_long <- pivot_longer(df_one,
cols=3:5,
names_to = "tissueType",
values_to = "Expression")
expression.df_two <- ggplot(df_two_long,
mapping = aes(y=tissueType, x=vars, fill = Expression))
geom_tile()
expression.df_two
CodePudding user response:
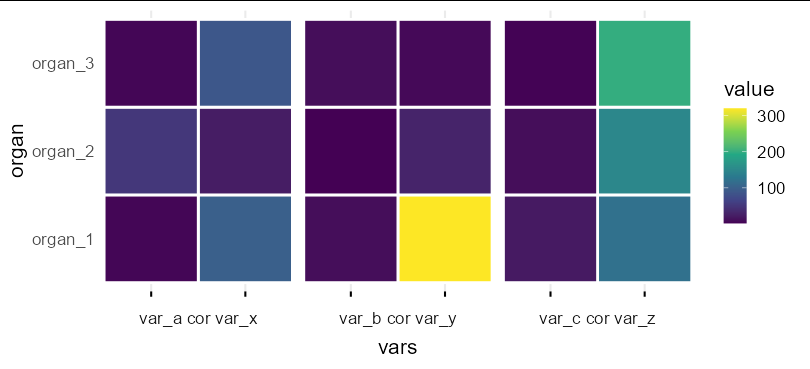
You can bind your data frames together and pivot into a longer format so that vars and corresponding vars are in the same column, but retain a grouping variable to facet by:
df_two %>%
mutate(cor = corresponding_vars) %>%
rename_with(~sub('corresponding_', '', .x)) %>%
bind_rows(df_one %>% rename(cor = corresponding_vars)) %>%
pivot_longer(contains('expression'), names_to = 'organ') %>%
mutate(organ = gsub('expression_|_vars', '', organ)) %>%
group_by(cor) %>%
summarize(vars = vars, organ = organ, value = value,
cor = paste(sort(unique(vars)), collapse = ' cor ')) %>%
ggplot(aes(vars, organ, fill = value))
geom_tile(color = 'white', linewidth = 1)
facet_grid(.~cor, scales = 'free_x', switch = 'x')
scale_fill_viridis_c()
coord_cartesian(clip = 'off')
scale_x_discrete(expand = c(0, 0))
theme_minimal(base_size = 16)
theme(strip.placement = 'outside',
axis.text.x = element_blank(),
axis.ticks.x.bottom = element_line(),
panel.spacing.x = unit(3, 'mm'))
CodePudding user response:
Okay, so I solved the issue for my own project, which is to convert it to a scatter plot. I combined both datasets and then used a simple scatterplot.
df.combined <- dplyr::full_join(df_two_long, df_one_long,
by = c("vars", "corresponding_vars", "tissueType"))
ggplot(df.combined,
aes(x=vars, y=tissueType, colour=Expression.x, size = Expression.y))
geom_point()
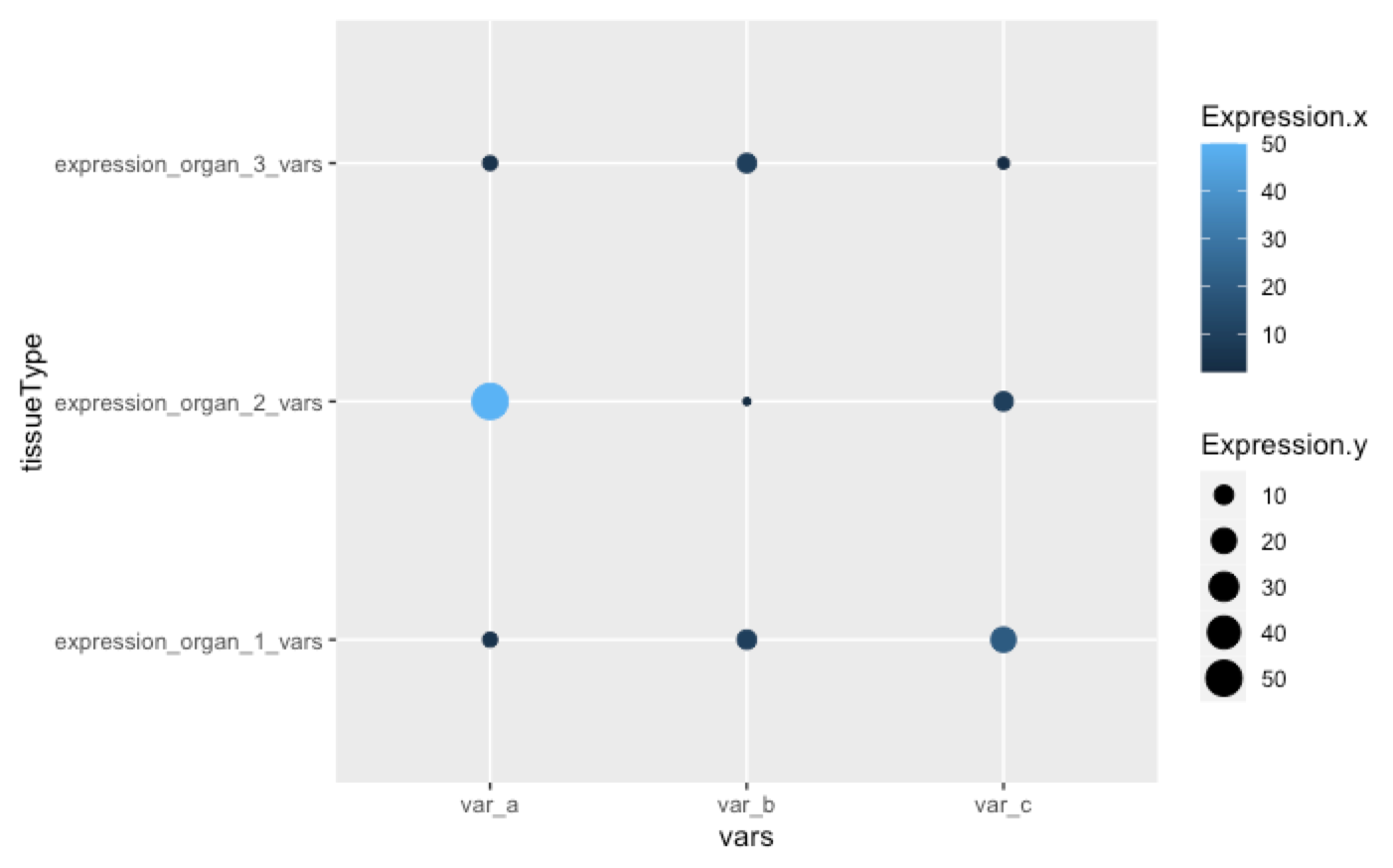 It's not a solution with heatmaps, but I don't know how to do that at the moment.
It's not a solution with heatmaps, but I don't know how to do that at the moment.