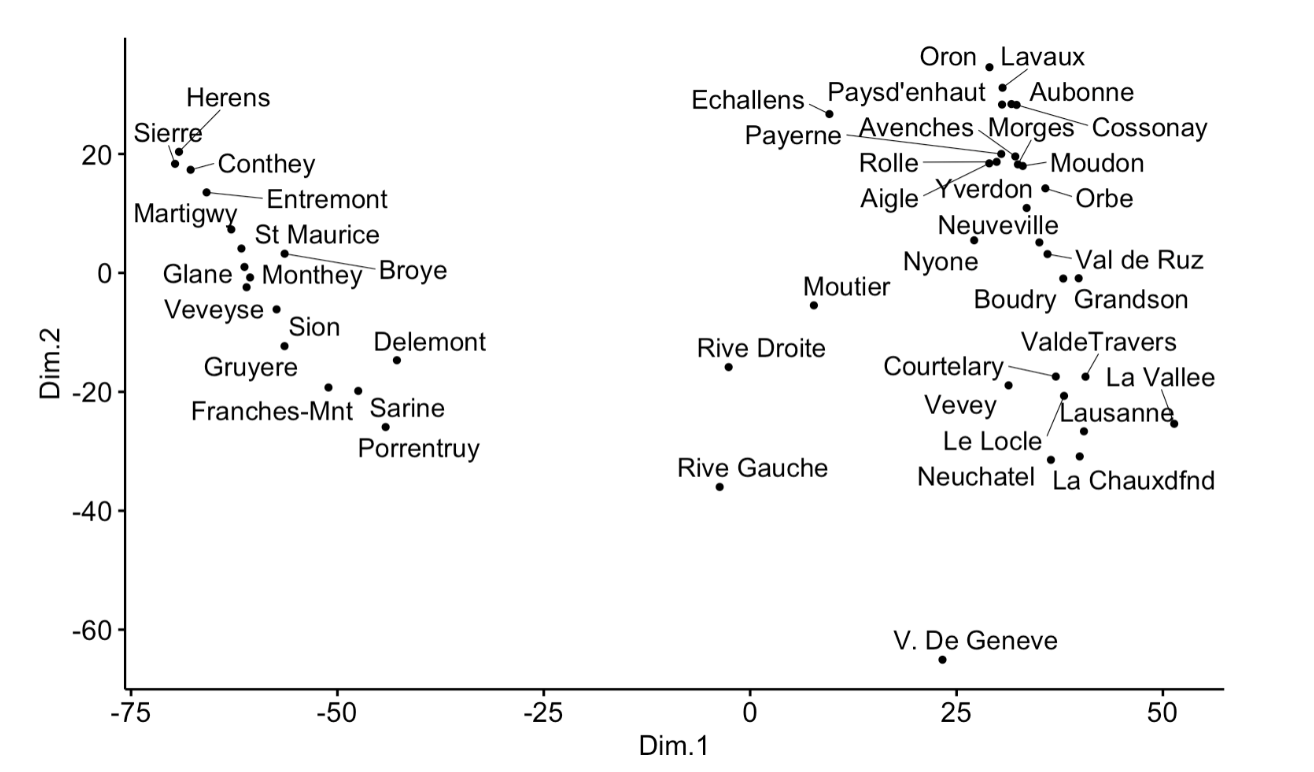
In MDS with R, how shall I specify the scale that I want to use? For example, I want Dim.1 to have a scale of -80 to 60, and Dim.2 to have a scale of -70 to 30?
data("swiss")
head(swiss)
library(magrittr)
library(dplyr)
library(ggpubr)
# Cmpute MDS
mds <- swiss %>%
dist() %>%
cmdscale() %>%
as_tibble()
colnames(mds) <- c("Dim.1", "Dim.2")
# Plot MDS
ggscatter(mds, x = "Dim.1", y = "Dim.2",
label = rownames(swiss),
size = 1,
repel = TRUE)
Source code taken from http://www.sthda.com/english/articles/31-principal-component-methods-in-r-practical-guide/122-multidimensional-scaling-essentials-algorithms-and-r-code/
CodePudding user response:
You can change your scales' breaks by adding other ggplot2 arguments.
data("swiss")
head(swiss)
#> Fertility Agriculture Examination Education Catholic
#> Courtelary 80.2 17.0 15 12 9.96
#> Delemont 83.1 45.1 6 9 84.84
#> Franches-Mnt 92.5 39.7 5 5 93.40
#> Moutier 85.8 36.5 12 7 33.77
#> Neuveville 76.9 43.5 17 15 5.16
#> Porrentruy 76.1 35.3 9 7 90.57
#> Infant.Mortality
#> Courtelary 22.2
#> Delemont 22.2
#> Franches-Mnt 20.2
#> Moutier 20.3
#> Neuveville 20.6
#> Porrentruy 26.6
library(magrittr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(ggpubr)
#> Loading required package: ggplot2
# Cmpute MDS
mds <- swiss %>%
dist() %>%
cmdscale() %>%
as_tibble()
#> Warning: The `x` argument of `as_tibble.matrix()` must have unique column names if
#> `.name_repair` is omitted as of tibble 2.0.0.
#> ℹ Using compatibility `.name_repair`.
colnames(mds) <- c("Dim.1", "Dim.2")
# Plot MDS
ggscatter(mds, x = "Dim.1", y = "Dim.2",
label = rownames(swiss),
size = 1,
repel = TRUE)
scale_y_continuous(breaks = seq(-70,30,20))
scale_x_continuous(breaks = seq(-80,60,20))
#> Warning: ggrepel: 6 unlabeled data points (too many overlaps). Consider
#> increasing max.overlaps
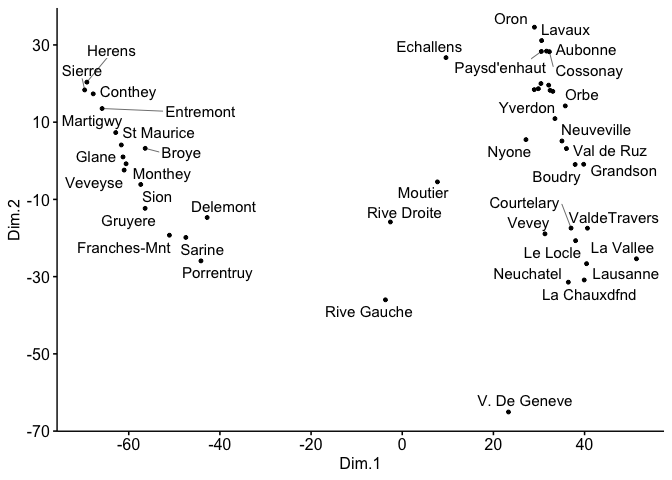
Created on 2023-01-07 with reprex v2.0.2
CodePudding user response:
Another option using coord_cartesian with expand = FALSE like this:
library(magrittr)
library(dplyr)
library(ggpubr)
# Plot MDS
ggscatter(mds, x = "Dim.1", y = "Dim.2",
label = rownames(swiss),
size = 1,
repel = TRUE)
coord_cartesian(xlim = c(-80,60), ylim = c(-70,30), expand = FALSE)
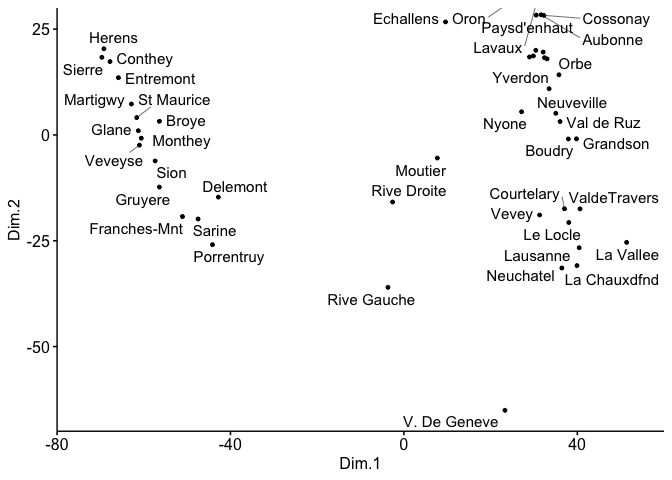
Created on 2023-01-07 with reprex v2.0.2