This table is based on a species sampling procedure that comprises starting at a certain location in a forest and recording the number of species that occur in that exact spot. Then, the surveyor walks and records the distance he traveled until he found a new species. This is the distance between the place where he found the new species and the initial point.
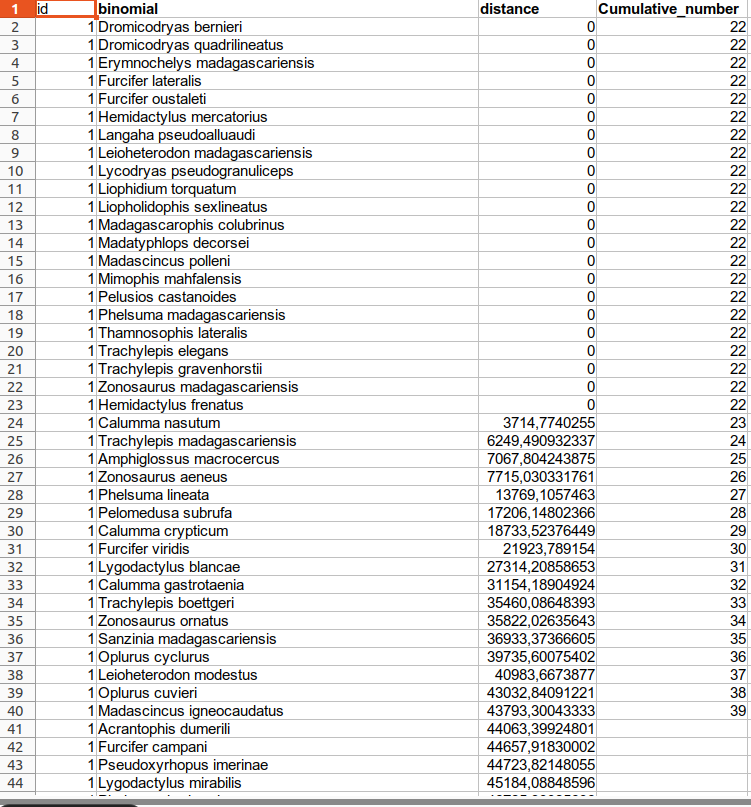
I would like to create a new column the includes the cumulative number of species based on the traveled distance. Here's what the new column should look like.
Example data:
data<-structure(list(id = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1), binomial = c("Dromicodryas bernieri",
"Dromicodryas quadrilineatus", "Erymnochelys madagascariensis",
"Furcifer lateralis", "Furcifer oustaleti", "Hemidactylus mercatorius",
"Langaha pseudoalluaudi", "Leioheterodon madagascariensis", "Lycodryas pseudogranuliceps",
"Liophidium torquatum", "Liopholidophis sexlineatus", "Madagascarophis colubrinus",
"Madatyphlops decorsei", "Madascincus polleni", "Mimophis mahfalensis",
"Pelusios castanoides", "Phelsuma madagascariensis", "Thamnosophis lateralis",
"Trachylepis elegans", "Trachylepis gravenhorstii", "Zonosaurus madagascariensis",
"Hemidactylus frenatus", "Calumma nasutum", "Trachylepis madagascariensis",
"Amphiglossus macrocercus", "Zonosaurus aeneus", "Phelsuma lineata",
"Pelomedusa subrufa", "Calumma crypticum", "Furcifer viridis",
"Lygodactylus blancae", "Calumma gastrotaenia", "Trachylepis boettgeri",
"Zonosaurus ornatus", "Sanzinia madagascariensis", "Oplurus cyclurus",
"Leioheterodon modestus", "Oplurus cuvieri", "Madascincus igneocaudatus",
"Acrantophis dumerili", "Furcifer campani", "Pseudoxyrhopus imerinae",
"Lygodactylus mirabilis", "Phelsuma barbouri", "Furcifer minor",
"Compsophis infralineatus", "Pseudoxyrhopus quinquelineatus",
"Calumma hilleniusi", "Paroedura bastardi", "Brookesia brygooi"
), distance = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 3714.77402549982, 6249.49093233716, 7067.80424387549,
7715.0303317613, 13769.1057463018, 17206.1480236598, 18733.5237644898,
21923.789153995, 27314.2085865309, 31154.1890492383, 35460.0864839256,
35822.0263564291, 36933.3736660544, 39735.6007540156, 40983.6673876956,
43032.8409122139, 43793.3004333338, 44063.3992480126, 44657.9183000201,
44723.8214805486, 45184.0884859559, 46785.9008560645, 48994.7048866502,
55332.621992021, 57746.4142325833, 58866.2845249788, 60839.811988087,
65560.1987963227)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-50L))
CodePudding user response:
One option is to get the cumsum on a logical vector i.e. where distance not equal to 0 and then add the count of 0 distance to it (sum(distance == 0))
library(dplyr)
data <- data %>%
mutate(new = cumsum(distance!=0) sum(distance == 0))
Or for this, we can use base R
data$new <- with(data, cumsum(distance!=0) sum(distance == 0))