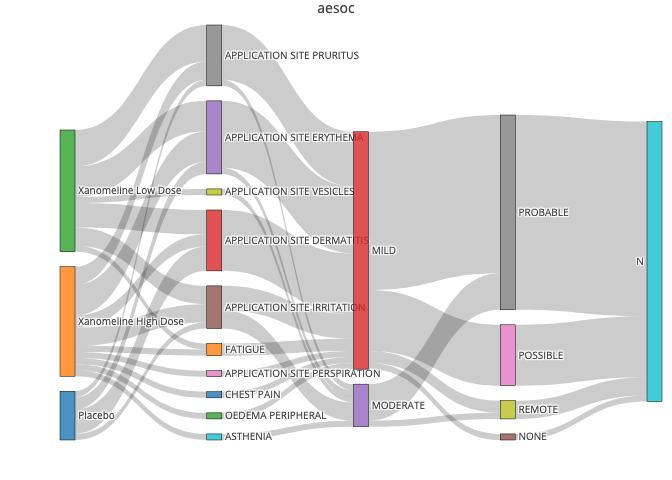
I tried to generate the sankey plot by plotly package, however there is a no error or warning but the graph is blank, could you please let me know the reason
I attached the data which I used, the data_long and nodes
I am not sure what is the reason, please help me with the reason and solution to generate the plot
data_long dataset
data_long <- structure(list(target = c("APPLICATION SITE DERMATITIS", "APPLICATION SITE DERMATITIS",
"APPLICATION SITE DERMATITIS", "APPLICATION SITE ERYTHEMA", "APPLICATION SITE ERYTHEMA",
"APPLICATION SITE ERYTHEMA", "APPLICATION SITE IRRITATION", "APPLICATION SITE IRRITATION",
"APPLICATION SITE IRRITATION", "APPLICATION SITE PERSPIRATION",
"APPLICATION SITE PRURITUS", "APPLICATION SITE PRURITUS", "APPLICATION SITE PRURITUS",
"APPLICATION SITE VESICLES", "ASTHENIA", "CHEST PAIN", "FATIGUE",
"FATIGUE", "OEDEMA PERIPHERAL", "MILD", "MILD", "MODERATE", "MILD",
"MODERATE", "MILD", "MILD", "MODERATE", "MODERATE", "MODERATE",
"MILD", "MILD", "MILD", "NONE", "POSSIBLE", "PROBABLE", "REMOTE",
"PROBABLE", "REMOTE", "N", "N", "N", "N"), source = c("Placebo",
"Xanomeline High Dose", "Xanomeline Low Dose", "Placebo", "Xanomeline High Dose",
"Xanomeline Low Dose", "Placebo", "Xanomeline High Dose", "Xanomeline Low Dose",
"Xanomeline High Dose", "Placebo", "Xanomeline High Dose", "Xanomeline Low Dose",
"Xanomeline Low Dose", "Xanomeline High Dose", "Xanomeline High Dose",
"Xanomeline High Dose", "Xanomeline Low Dose", "Xanomeline High Dose",
"APPLICATION SITE DERMATITIS", "APPLICATION SITE ERYTHEMA", "APPLICATION SITE ERYTHEMA",
"APPLICATION SITE IRRITATION", "APPLICATION SITE IRRITATION",
"APPLICATION SITE PERSPIRATION", "APPLICATION SITE PRURITUS",
"APPLICATION SITE PRURITUS", "APPLICATION SITE VESICLES", "ASTHENIA",
"CHEST PAIN", "FATIGUE", "OEDEMA PERIPHERAL", "MILD", "MILD",
"MILD", "MILD", "MODERATE", "MODERATE", "NONE", "POSSIBLE", "PROBABLE",
"REMOTE"), value = c(4L, 2L, 4L, 2L, 5L, 5L, 1L, 3L, 3L, 1L,
1L, 3L, 6L, 1L, 1L, 1L, 1L, 1L, 1L, 10L, 11L, 1L, 4L, 3L, 1L,
9L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 10L, 26L, 2L, 6L, 1L, 1L, 10L,
32L, 3L), IDsource = c(0, 1, 2, 0, 1, 2, 0, 1, 2, 1, 0, 1, 2,
2, 1, 1, 1, 2, 1, 3, 4, 4, 5, 5, 6, 7, 7, 8, 9, 10, 11, 12, 13,
13, 13, 13, 14, 14, 15, 16, 17, 18), IDtarget = c(3, 3, 3, 4,
4, 4, 5, 5, 5, 6, 7, 7, 7, 8, 9, 10, 11, 11, 12, 13, 13, 14,
13, 14, 13, 13, 14, 14, 14, 13, 13, 13, 15, 16, 17, 18, 17, 18,
19, 19, 19, 19)), row.names = c(NA, -42L), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), groups = structure(list(target = c("APPLICATION SITE DERMATITIS",
"APPLICATION SITE DERMATITIS", "APPLICATION SITE DERMATITIS",
"APPLICATION SITE ERYTHEMA", "APPLICATION SITE ERYTHEMA", "APPLICATION SITE ERYTHEMA",
"APPLICATION SITE IRRITATION", "APPLICATION SITE IRRITATION",
"APPLICATION SITE IRRITATION", "APPLICATION SITE PERSPIRATION",
"APPLICATION SITE PRURITUS", "APPLICATION SITE PRURITUS", "APPLICATION SITE PRURITUS",
"APPLICATION SITE VESICLES", "ASTHENIA", "CHEST PAIN", "FATIGUE",
"FATIGUE", "MILD", "MILD", "MILD", "MILD", "MILD", "MILD", "MILD",
"MILD", "MODERATE", "MODERATE", "MODERATE", "MODERATE", "MODERATE",
"N", "N", "N", "N", "NONE", "OEDEMA PERIPHERAL", "POSSIBLE",
"PROBABLE", "PROBABLE", "REMOTE", "REMOTE"), source = c("Placebo",
"Xanomeline High Dose", "Xanomeline Low Dose", "Placebo", "Xanomeline High Dose",
"Xanomeline Low Dose", "Placebo", "Xanomeline High Dose", "Xanomeline Low Dose",
"Xanomeline High Dose", "Placebo", "Xanomeline High Dose", "Xanomeline Low Dose",
"Xanomeline Low Dose", "Xanomeline High Dose", "Xanomeline High Dose",
"Xanomeline High Dose", "Xanomeline Low Dose", "APPLICATION SITE DERMATITIS",
"APPLICATION SITE ERYTHEMA", "APPLICATION SITE IRRITATION", "APPLICATION SITE PERSPIRATION",
"APPLICATION SITE PRURITUS", "CHEST PAIN", "FATIGUE", "OEDEMA PERIPHERAL",
"APPLICATION SITE ERYTHEMA", "APPLICATION SITE IRRITATION", "APPLICATION SITE PRURITUS",
"APPLICATION SITE VESICLES", "ASTHENIA", "NONE", "POSSIBLE",
"PROBABLE", "REMOTE", "MILD", "Xanomeline High Dose", "MILD",
"MILD", "MODERATE", "MILD", "MODERATE"), .rows = structure(list(
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L,
15L, 16L, 17L, 18L, 20L, 21L, 23L, 25L, 26L, 30L, 31L, 32L,
22L, 24L, 27L, 28L, 29L, 39L, 40L, 41L, 42L, 33L, 19L, 34L,
35L, 37L, 36L, 38L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -42L), .drop = TRUE))
nodes dataset
nodes <- structure(list(name = c("Placebo", "Xanomeline High Dose", "Xanomeline Low Dose",
"APPLICATION SITE DERMATITIS", "APPLICATION SITE ERYTHEMA", "APPLICATION SITE IRRITATION",
"APPLICATION SITE PERSPIRATION", "APPLICATION SITE PRURITUS",
"APPLICATION SITE VESICLES", "ASTHENIA", "CHEST PAIN", "FATIGUE",
"OEDEMA PERIPHERAL", "MILD", "MODERATE", "NONE", "POSSIBLE",
"PROBABLE", "REMOTE", "N")), class = "data.frame", row.names = c(NA,
-20L))
#plotly
fig <- plot_ly(
type = "sankey",
domain = list(
x = c(0,1),
y = c(0,1)
),
orientation = "h",
valueformat = ".0f",
valuesuffix = "TWh",
node = list(
label = nodes$name,
pad = 15,
thickness = 15,
line = list(
color = "black",
width = 0.5
)
),
link = list(
source = data_long$source,
target = data_long$target,
value = data_long$value
)
)
fig <- fig %>% layout(
title = "aesoc",
font = list(
size = 10
),
xaxis = list(showgrid = F, zeroline = F),
yaxis = list(showgrid = F, zeroline = F)
)
fig
CodePudding user response:
You have to use the source and target ID columns from your dataset:
library(plotly)
fig <- plot_ly(
type = "sankey",
domain = list(
x = c(0, 1),
y = c(0, 1)
),
orientation = "h",
valueformat = ".0f",
valuesuffix = "TWh",
node = list(
label = nodes$name,
pad = 15,
thickness = 15,
line = list(
color = "black",
width = 0.5
)
),
link = list(
source = data_long$IDsource,
target = data_long$IDtarget,
value = data_long$value
)
)
fig %>% layout(
title = "aesoc",
font = list(
size = 10
),
xaxis = list(showgrid = F, zeroline = F),
yaxis = list(showgrid = F, zeroline = F)
)