I want to add a predetermined correlation between two skew-normal variables generated with fGarch::rsnorm(). The variables also need to be positive with a predetermined mean, variance, and skewness.
Here is my attempt:
correlation <- 0.3
mean_1 <- 20
mean_2 <- 10
sd_1 <- 10
sd_2 <- 10
xi_1 <- 10
xi_2 <- 10
n <- 10000
x <- abs(fGarch::rsnorm(n, mean = mean_1, sd = sd_1, xi = xi_1))
y <- abs(fGarch::rsnorm(n, mean = mean_2, sd = sd_2, xi = xi_2))
x <- abs(x correlation * (y - mean(y)))
y <- abs(y correlation * (x - mean(x)))
hist(x)
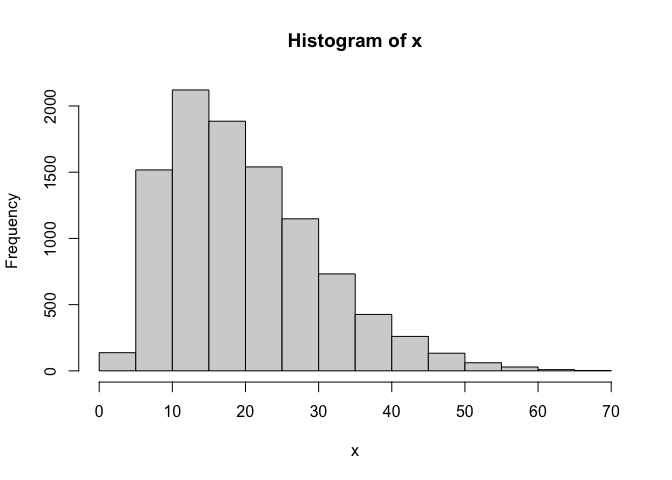
hist(y)
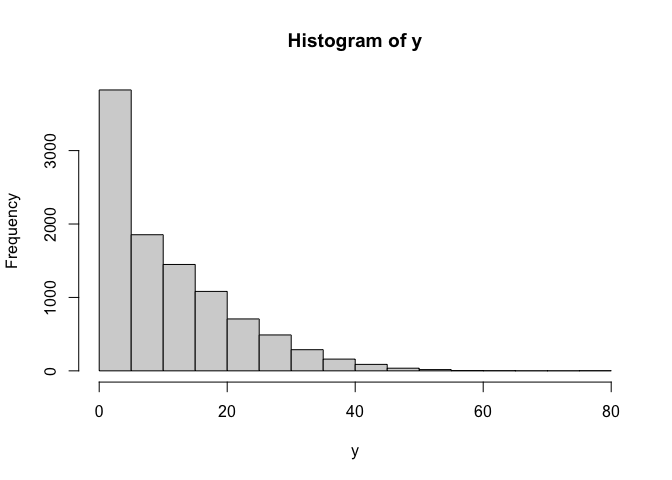
cor(x,y)
#> [1] 0.4941751
Created on 2023-01-13 with reprex v2.0.2
CodePudding user response:
Let's recreate your example but with a reproducible random seed
set.seed(1)
correlation <- 0.3
mean_1 <- 20
mean_2 <- 10
sd_1 <- 10
sd_2 <- 10
xi_1 <- 10
xi_2 <- 10
n <- 10000
x <- abs(fGarch::rsnorm(n, mean = mean_1, sd = sd_1, xi = xi_1))
y <- abs(fGarch::rsnorm(n, mean = mean_2, sd = sd_2, xi = xi_2))
Now of course x and y are effectively completely uncorrelated:
cor(x, y)
#> [1] -0.003209725
But if we sort both vectors, they become almost perfectly correlated.
x <- sort(x)
y <- sort(y)
cor(x, y)
#> [1] 0.9967296
But we want something in between, so let's shuffle x randomly, one pair at a time, and keep doing this until the correlation just falls below our target correlation:
while(cor(x, y) > correlation) {
ij <- sample(n, 2)
tmpx <- x[ij[1]]
x[ij[1]] <- x[ij[2]]
x[ij[2]] <- tmpx
}
Of course, we haven't changed the actual value of x or y, so their parameters are unchanged:
hist(x)
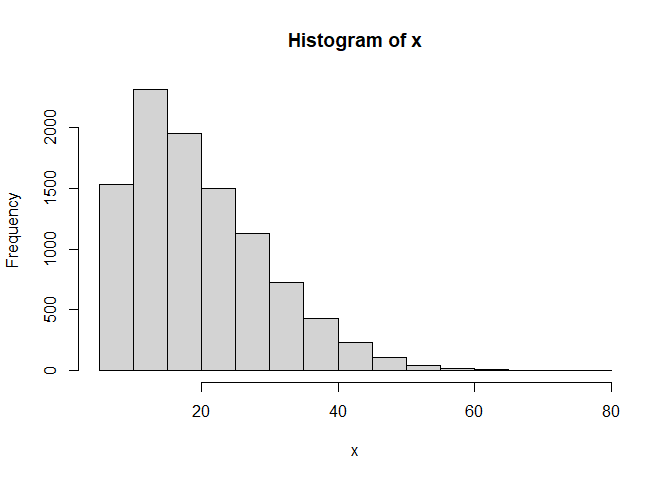
hist(y)
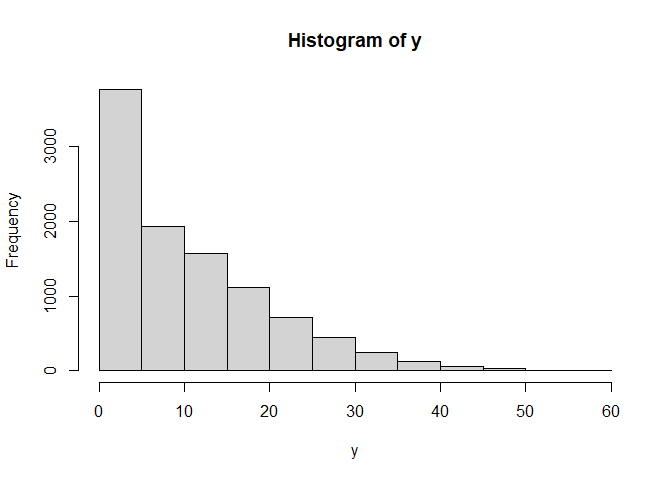
mean(x)
#> [1] 19.87497
mean(y)
#> [1] 10.60162
But now their correlation matches our target value:
cor(x, y)
#> [1] 0.2997019
Created on 2023-01-13 with reprex v2.0.2
CodePudding user response:
Here's a starting point: to get this to work properly I'd have to spend more time reading the documentation and the relevant papers to figure out what the parameterizations are.
Start with library("sos"); findFn("{multivariate skew-normal}") to find the sn package. Then look at the examples in ?rmsn. This leads us to ?cp2dp:
For a multivariate distribution, there exists an extension based on the same logic; its components represent the vector mean value, the variance matrix, the vector of marginal coefficients of skewness ...
mean_v <- c(20, 10)
sd_v <- c(10, 10)
correlation <- 0.3
## from SD, cor to covariance matrix
vv <- outer(sd_v, sd_v) * matrix(c(1, correlation, correlation, 1), 2, 2)
xi_v <- c(10, 10)
n <- 10000
## note different notation in sn vs fGarch: xi is location
## n.b. I haven't checked that sn 'alpha' is actually
## equivalent to fGarch 'xi' ...
op_pars <- list(xi=mean_v, Omega=vv, alpha=xi_v)
dp_pars <- op2dp(op_pars, family = "SN")
set.seed(101)
y <- rmsn(n = n, dp = dp_pars)
colMeans(y)
