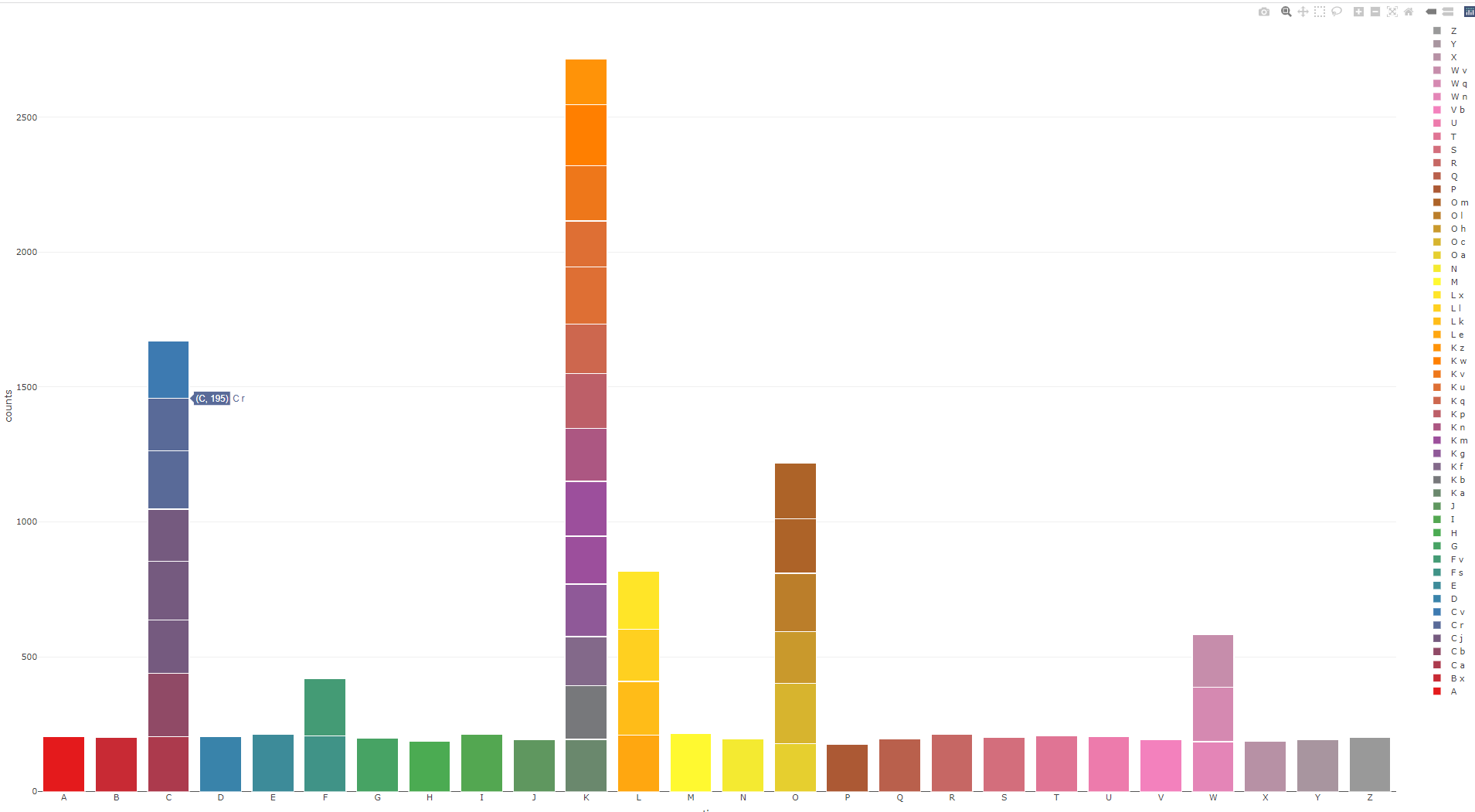
Im trying to replicate a barplot (See picture below) in a Shiny app using Plotly in R.
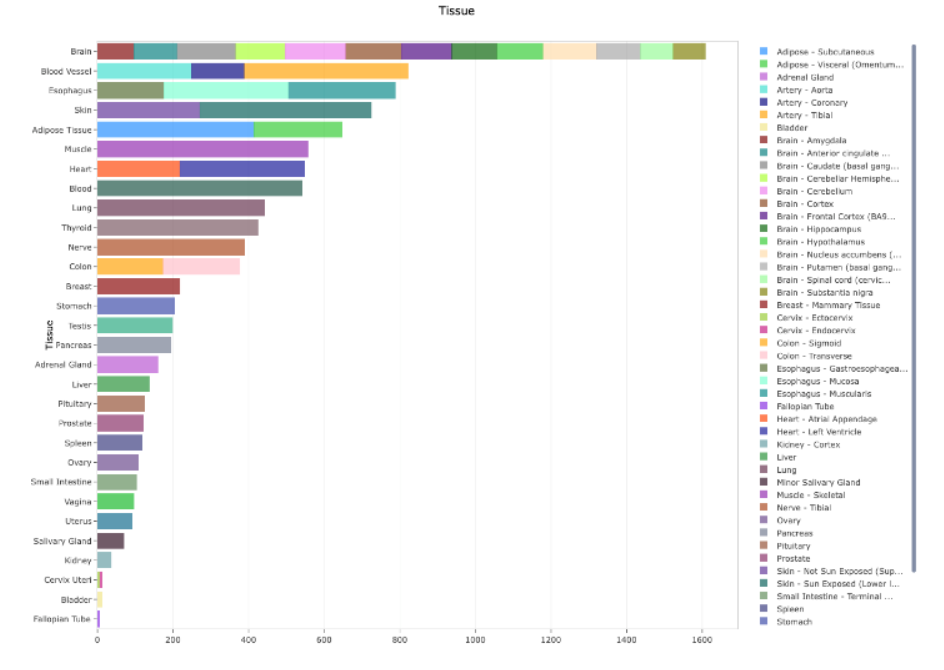 The x-axis has tissue values, while each bar on the x-axis is split/stacked based on tissue details.
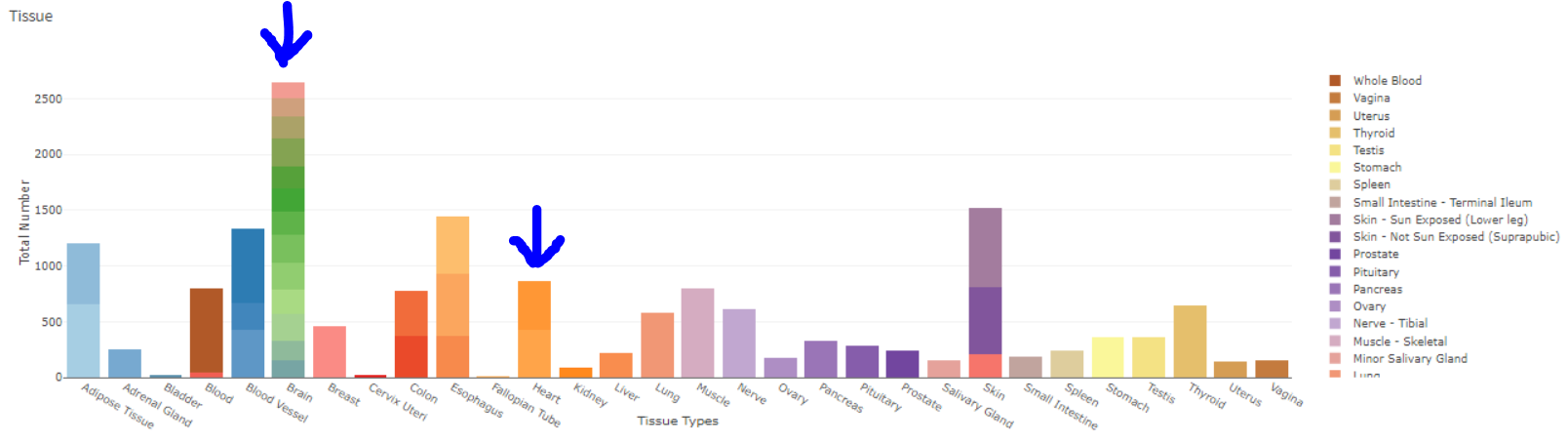
When I try to replicate the above graph, For some tissue values, I am unable to discretely differentiate between tissue details using the predefined plotly color scales (See 'Brain' and 'heart' below), The bars basically have variations of the same color family, making them hard to interpret.
The x-axis has tissue values, while each bar on the x-axis is split/stacked based on tissue details.
When I try to replicate the above graph, For some tissue values, I am unable to discretely differentiate between tissue details using the predefined plotly color scales (See 'Brain' and 'heart' below), The bars basically have variations of the same color family, making them hard to interpret.
Is it possible somehow to produce a graph with colors like the one I want to replicate, in plotly R? Dynamically? As the tissue details for a tissue could change based on shiny select input selections.
This is my plotly function:
plot_ly(data = tissue_dt, y=~counts, x=tissue, type='bar', name=~tissue_detail, color=~tissue_detail, colors = "Paired") %>% layout(barmode ="stack")
Apart from this I have also tried "Set3" and all other plotly discrete colorscales, but to no luck.
The data being used for both the desired plot and the shiny plot, is the same
CodePudding user response:
With so many (sub) categories it will be rather difficult to define a meaningful discrete palette. However, with plotly's hover capabilities and by adding a (white) line around the bars, you should get a farily decent graph:
library(dplyr)
library(tibble)
library(plotly)
library(RColorBrewer)
tissues <- tibble(tissue = LETTERS)
set.seed(18012023)
tissue_dt <- tissues %>%
slice(rep(sample(26, 8), sample(14, 8, TRUE))) %>%
mutate(tissue_detail = paste(tissue, sample(letters, n(), TRUE))) %>%
right_join(tissues, "tissue") %>%
mutate(tissue_detail = coalesce(tissue_detail, tissue),
counts = rpois(n(), 200)) %>%
arrange(tissue)
plot_ly(data = tissue_dt, y = ~counts, x = ~ tissue,
type = "bar",
name = ~tissue_detail,
marker = list(line = list(color = "white",
width = 1.5)),
color = ~tissue_detail,
colors = ~ tissue_detail %>%
as.factor() %>%
nlevels() %>%
colorRampPalette(brewer.pal(9, "Set1"))(.)) %>%
layout(barmode ="stack")
CodePudding user response:
Note that the original plot uses a discrete color for each tissue/detail.
The best option, if the list of the tissue_detail is fixed is to use a manual palette like:
my.pal = {
"Adipose-subcutaneous" = "#a10335", #Or whatever color you want
"Adipose-visceral" = "#43a332",
"AdrenalGland" = "green2",
# AND the remaining categories. The labels should match those in tissue_detail
}
And the just use that palette in your plot
plot_ly(data = tissue_dt, y=~counts, x=tissue, type='bar', name=~tissue_detail, color=~tissue_detail, colors = my.pal) %>% layout(barmode ="stack")
This have an additional advantage if you use these data partially in other plots. Thus you can keep the assignation of colors to each tissue_detail even if there is some missing category in any of those plots.