I'm wondering how different dtype of numpy arrays affect the memory usage in Python when calling .tolist() on the array. I want to conserve as much memory as possible. If I am correct, using float16 shoudl allow me to carry double the amount of values when compared to float32 because float16 uses half as much memory as float32 but the thing is I don't seem to see a difference in the byte size when I call .tolist() onto the numpy array. Does Python not conserve the dtype and just revert everything to the exact same dtype or am I missing something in my observations?
Example:
import xarray as xr
import numpy as np
import sys
x = xr.DataArray(np.arange(6).reshape(2,3),dims=["lat","lon"],coords={"lat":[1,2], "lon": [0,1,2]})
m = xr.zeros_like(x, dtype= 'float64')
m2 = xr.zeros_like(x, dtype= 'float32')
m3 = xr.zeros_like(x, dtype= 'float16')
print("m")
print(m.dtype)
print(m.nbytes)
print(sys.getsizeof(m))
print(sys.getsizeof(m.values.tolist()))
print("m2")
print(m2.dtype)
print(m2.nbytes)
print(sys.getsizeof(m2))
print(sys.getsizeof(m2.values.tolist()))
print("m3")
print(m3.dtype)
print(m3.nbytes)
print(sys.getsizeof(m3))
print(sys.getsizeof(m3.values.tolist()))
The output is the follwoing:
m
float64
48
56
80
m2
float32
24
56
80
m3
float16
12
56
80
It states that calling .tolist() uses 80 bytes regardless of dtype. So my question is, is there even any benefit to memeory usage in python when converting numpy array to list in python. Is there ANY memory benefit at all at ANY point in the process of m3.values.tolist() with regards to using a different dtype for the numpy data or is there no point in trying to conserve memory usage and am I just lowering precision using float16
CodePudding user response:
In [39]: x = xr.DataArray(np.arange(6).reshape(2,3),dims=["lat","lon"],coords={"lat":[1,2], "lon": [0,1,2]})
...:
...: m = xr.zeros_like(x, dtype= 'float64')
...: m2 = xr.zeros_like(x, dtype= 'float32')
...: m3 = xr.zeros_like(x, dtype= 'float16')
In [40]: m.shape
Out[40]: (2, 3)
Because m is 2d, the list equivalent is a list of lists. Keep that in mind when considering memory use.
In [41]: m.values.tolist()
Out[41]: [[0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]
First check the type of individual numeric elements - all the same:
In [42]: type(m.values.tolist()[0][0])
Out[42]: float
In [43]: type(m2.values.tolist()[0][0])
Out[43]: float
In [44]: type(m3.values.tolist()[0][0])
Out[44]: float
As for getsizeof applied to list - it just measures the memory allocated for storing references (plus some overhead):
In [45]: sys.getsizeof(m.values.tolist()) # a len 2 list
Out[45]: 72
In [46]: sys.getsizeof(m.values.tolist()[0]) # a len 3 list (on more pointer)
Out[46]: 80
An individual float:
In [47]: sys.getsizeof(m.values.tolist()[0][0])
Out[47]: 24
So total size will be 72 280 624
Since your goal is a json string (or file) let's try encoding:
In [48]: import json
In [50]: json.dumps(m.values.tolist())
Out[50]: '[[0.0, 0.0, 0.0], [0.0, 0.0, 0.0]]'
But for random float values:
In [55]: json.dumps(np.random.randn(2,3).tolist())
Out[55]: '[[0.8431542062798922, 0.939582071772101, 0.8783338312384453], [-0.5734061911788975, -0.3403280836021266, 0.39779755792940164]]'
Fiddling with the dtype of the source array changes nothing.
CodePudding user response:
Calling tolist returns a pure-python object, so it returns a list of floats. Python has no distinction of different sizes of floats -- they are all of the same type.
m_l = m.tolist()
m2_l = m2.tolist()
m3_l = m3.tolist()
print(type(m_l[0][0])) # <class 'float'>
print(type(m2_l[0][0])) # <class 'float'>
print(type(m3_l[0][0])) # <class 'float'>
print(type(m_l[0][0]) == type(m2_l[0][0]) == type(m3_l[0][0]))
# True
is there even any benefit to memory usage in python when converting numpy array to list in python. Is there ANY memory benefit at all at ANY point in the process of
m3.values.tolist()with regards to using a different dtype for the numpy data or is there no point in trying to conserve memory usage and am I just lowering precision using float16
Here's a comparison of runtimes for functions that sum an array by
- Iterating over
arr.tolist() - Iterating over
arrdirectly - Calling the numpy method
arr.sum()
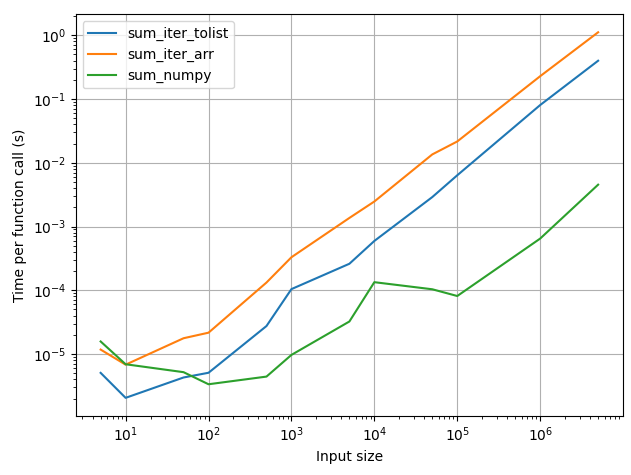
While iterating over arr.tolist() is faster than simply iterating over the array, calling arr.sum() is faster for large arrays. If you must iterate over the array, you could do it using .tolist, but as far as possible you should try to do it within numpy.
The choice of dtype depends on what you're using the array for. If you're using it for lat/lon, think about what is the max value it can be? Since calculations between float16 and float16 will yield another float16, can you do calculations in float16 precision?
Timing code:
import timeit
import numpy as np
from matplotlib import pyplot as plt
def time_funcs(funcs, sizes, arg_gen, N=20):
times = np.zeros((len(sizes), len(funcs)))
gdict = globals().copy()
for i, s in enumerate(sizes):
args = arg_gen(s)
print(args)
for j, f in enumerate(funcs):
gdict.update(locals())
try:
times[i, j] = timeit.timeit("f(*args)", globals=gdict, number=N) / N
print(f"{i}/{len(sizes)}, {j}/{len(funcs)}, {times[i, j]}")
except ValueError:
print("ERROR in {f}({*args})")
return times
def plot_times(times, funcs):
fig, ax = plt.subplots()
for j, f in enumerate(funcs):
ax.plot(sizes, times[:, j], label=f.__name__)
ax.set_xlabel("Array size")
ax.set_ylabel("Time per function call (s)")
ax.set_xscale("log")
ax.set_yscale("log")
ax.legend()
ax.grid()
fig.tight_layout()
return fig, ax
#%%
def arg_gen(n):
return [np.random.random((n,))]
#%%
def sum_iter_tolist(arr):
lst = arr.tolist()
x = 0
for i in lst:
x = i
return x
def sum_iter_arr(arr):
x = 0
for i in arr:
x = i
return x
def sum_numpy(arr):
return np.sum(arr)
#%%
if __name__ == "__main__":
#%% Set up sim
# sizes = [5, 10, 50, 100, 500, 1000, 5000, 10_000, 50_000, 100_000]
sizes = [5, 10, 50, 100, 500, 1000, 5000, 10_000, 50_000, 100_000, 1_000_000, 5_000_000]
funcs = [sum_iter_tolist, sum_iter_arr, sum_numpy]
#%% Run timing
time_fcalls = np.zeros((len(sizes), len(funcs))) * np.nan
time_fcalls = time_funcs(funcs, sizes, arg_gen)
#%%
fig, ax = plot_times(time_fcalls, funcs)
ax.set_xlabel(f"Input size")
plt.show()
