I run the same test code in Python Numpy and in Matlab and see that the Matlab code is faster by an order of magnitude. I want to know what is the bottleneck of the Python code and how to speed it up.
I run the following test code using Python Numpy (the last part is the performance sensitive one):
# Packages
import numpy as np
import time
# Number of possible outcomes
num_outcomes = 20
# Dimension of the system
dim = 50
# Number of iterations
num_iterations = int(1e7)
# Possible outcomes
outcomes = np.arange(num_outcomes)
# Possible transition matrices
matrices = [np.random.rand(dim, dim) for k in outcomes]
matrices = [mat/np.sum(mat, axis=0) for mat in matrices]
# Initial state
state = np.random.rand(dim)
state = state/np.sum(state)
# List of samples
samples = np.random.choice(outcomes, size=(num_iterations,))
samples = samples.tolist()
# === PERFORMANCE-SENSITIVE PART OF THE CODE ===
# Update the state over all iterations
start_time = time.time()
for k in range(num_iterations):
sample = samples[k]
matrix = matrices[sample]
state = np.matmul(matrix, state)
end_time = time.time()
# Print the execution time
print(end_time - start_time)
I then run an equivalent code using Matlab (the last part is the performance sensitive one):
% Number of possible outcomes
num_outcomes = 20;
% Number of dimensions
dim = 50;
% Number of iterations
num_iterations = 1e7;
% Possible outcomes
outcomes = 1:num_outcomes;
% Possible transition matrices
matrices = rand(num_outcomes, dim, dim);
matrices = matrices./sum(matrices,2);
matrices = num2cell(matrices,[2,3]);
matrices = cellfun(@shiftdim, matrices, 'UniformOutput', false);
% Initial state
state = rand(dim,1);
state = state./sum(state);
% List of samples
samples = datasample(outcomes, num_iterations);
% === PERFORMANCE-SENSITIVE PART OF THE CODE ===
% Update the state over all iterations
tic;
for k = 1:num_iterations
sample = samples(k);
matrix = matrices{sample};
state = matrix * state;
end
toc;
The Python code is consistently slower than the Matlab code by an order of magnitude, and I am not sure why.
Any idea where to start?
I run the Python code with the Python 3.10 interpreter and Numpy 1.22.4. I run the Matlab code with Matlab R2022a. Both codes are run on Windows 11 Pro 64 bits on a Lenovo T14 ThinkPad with the following processors:
11th Gen Intel(R) Core(TM) i7-1165G7 @ 2.80GHz, 2803 Mhz, 4 Core(s), 8 Logical Processor(s)
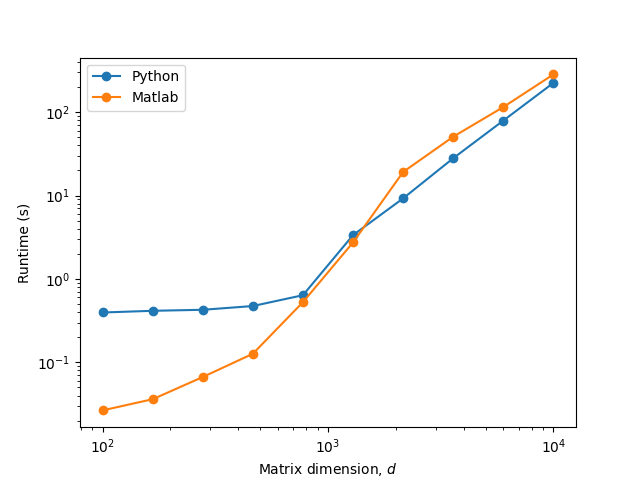
EDIT 1: I made some additional tests and it looks like the culprit is some type of Python-specific constant overhead at low matrix sizes:
As hpaulj and MSS suggest, this might mean that a JIT compiler could solve some of these issues. I will do my best to try this in the near future.
CodePudding user response:
In the loop, the main hindrance is np.matmul(matrix,state).
If we unroll the loop:
st[1] = m[0]@st[0]
st[2] = m[1]@st[1] = m[1]@m[0]@st[0]
st[3] = m[2]@m[1]@m[0]@st[0]
There is no obvious vectorized way to do looped np.matmul in a non loopy way.
A better way would be to do it in log_2(n) loops.
import numpy as np
outcomes = 20
dim = 50
num_iter = int(1e7)
mat = np.random.rand(outcomes,dim, dim)
mat = mat/mat.sum(axis=1)[...,None]
state = np.random.rand(dim)
state = state/np.sum(state)
samples = np.random.choice(np.arange(outcomes), size=(num_iter,))
a = mat[samples,...]
# This while loop takes log_2(num_iter) iterations
while len(a) > 1:
a = np.matmul(a[::2, ...], a[1::2, ...])
state = np.matmul(a,state)
The time may be further reduced by using numba jit.