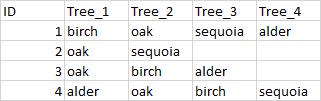
I have a dataset in which observation of one event occurs in different variables. To make a more clear example, imagine this: 4 people are told to go out in the woods and register trees. They are told to note the type of tree in the order they stumble upon them. Hence, each person hands in a list of the 1st to the 4th tree they found. It results in this data frame:
treedata <- structure(list(ID = c(1, 2, 3, 4), Tree_1 = c("birch", "oak",
"oak", "alder"), Tree_2 = c("oak", "sequoia", "birch", "oak"),
Tree_3 = c("sequoia", NA, "alder", "birch"), Tree_4 = c("alder",
NA, NA, "sequoia")), class = "data.frame", row.names = c(NA,
4L))
And the data looks like this
The data scientist is then told to count the number of observation for each tree type. But the problem is that for example "birch" appears in the variable "Tree_1" for ID 1, "Tree_2 for ID 3 and "Tree_3" for ID 4.
Basically what I want to do here is transform the Tree_x variables to a "birch" variable, "oak" variable and so forth and then assign a value of Yes or No if the ID stumbled upon that tree. Besides counting the trees, the new variables will be used to correlate the trees to a numerical variable.
My first idea was to use "unite", then rearrange so that each tree comes in the same order, then create a new variable. However I did not succeed in this and since there are NAs it proved a bit difficult still.
Does anyone have a solution? I tried googling and searching SO without any luck.
CodePudding user response:
Using recast
library(reshape2)
recast(treedata, id.var = 'ID', ID ~ value,
\(x) c('', 'yes')[1 (length(x) > 0)])[-6]
-output
ID alder birch oak sequoia
1 1 yes yes yes yes
2 2 yes yes
3 3 yes yes yes
4 4 yes yes yes yes
CodePudding user response:
If you're able to use the tidyverse packages, the first step is to transform the data to a long format that collapses the Tree columns:
library(tidyverse)
treedata_long <- pivot_longer(treedata, -ID, names_to = 'tree_num', values_to = 'tree_name') %>%
filter(!is.na(tree_name))
ID tree_num tree_name
<dbl> <chr> <chr>
1 1 Tree_1 birch
2 1 Tree_2 oak
3 1 Tree_3 sequoia
4 1 Tree_4 alder
5 2 Tree_1 oak
6 2 Tree_2 sequoia
7 3 Tree_1 oak
8 3 Tree_2 birch
9 3 Tree_3 alder
10 4 Tree_1 alder
11 4 Tree_2 oak
12 4 Tree_3 birch
13 4 Tree_4 sequoia
Then it's simple to use the count() function:
tree_counts <- count(treedata_long, tree_name)
tree_name n
<chr> <int>
1 alder 3
2 birch 3
3 oak 4
4 sequoia 3
We can then use pivot_wider to create the presence/absence matrix in your desired output:
tree_wide <- treedata_long %>%
select(-tree_num) %>%
pivot_wider(names_from = tree_name, values_from = tree_name, values_fn = \(x) ifelse(!is.na(x), 'yes', NA))
ID birch oak sequoia alder
<dbl> <chr> <chr> <chr> <chr>
1 1 yes yes yes yes
2 2 NA yes yes NA
3 3 yes yes NA yes
4 4 yes yes yes yes