I am trying to read in a list of shapefiles from a list of zip archives without actually unzipping the archives. Yes, I know that the archives will be unzipped in the background, but what I want to avoid is seeing the unzipped files in Windows Explorer.
This example can be fully reproducible, you need to download all the files from this Github repository and set your working directory to the folder where you downloaded them.
I also want to do it tidyverse-style, with pipes and without saving intermediate objects.
The code that I am currently trying to run is this one:
library(tidyverse)
library(magrittr)
library(sf)
list.files() %>%
map(unzip, list = T) %>%
map(filter, grepl(".shp$", Name)) %>%
map(~ .x %$% Name) %>%
map2(.x = ., .y = list.files(), .f = ~st_read(unzip(zipfile = .y, files = .x)))
However, that doesn't work. Why?
EDIT: To make the example more minimal, I guess you could also download just two of the files from the above repository.
CodePudding user response:
/vsizip GDAL Virtual File System driver is kind of convenient:
library(sf)
#> Linking to GEOS 3.9.3, GDAL 3.5.2, PROJ 8.2.1; sf_use_s2() is TRUE
library(dplyr)
library(stringr)
library(purrr)
(file_list <- list.files(pattern = "\\.zip$"))
#> [1] "tl_2019_01_place.zip" "tl_2019_02_place.zip"
sf_list <- file_list %>%
# resulting list will have names without ".zip"
set_names(str_remove(.,"\\.zip$")) %>%
map( ~ st_read(paste0("/vsizip/", .x)))
#> Reading layer `tl_2019_01_place' from data source `/vsizip/tl_2019_01_place.zip' using driver `ESRI Shapefile'
#> Simple feature collection with 586 features and 16 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -88.4442 ymin: 30.19825 xmax: -84.96303 ymax: 34.99807
#> Geodetic CRS: NAD83
#> Reading layer `tl_2019_02_place' from data source `/vsizip/tl_2019_02_place.zip' using driver `ESRI Shapefile'
#> Simple feature collection with 354 features and 16 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -176.6967 ymin: 51.81049 xmax: 173.4299 ymax: 71.34019
#> Geodetic CRS: NAD83
# 1st sf in the list:
sf_list$tl_2019_01_place %>% select(NAME, geometry)
#> Simple feature collection with 586 features and 1 field
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -88.4442 ymin: 30.19825 xmax: -84.96303 ymax: 34.99807
#> Geodetic CRS: NAD83
#> First 10 features:
#> NAME geometry
#> 1 Berry MULTIPOLYGON (((-87.6391 33...
#> 2 Fayette MULTIPOLYGON (((-87.85507 3...
#> 3 Gu-Win MULTIPOLYGON (((-87.88578 3...
#> 4 Ashville MULTIPOLYGON (((-86.30442 3...
#> 5 Margaret MULTIPOLYGON (((-86.46153 3...
#> 6 Odenville MULTIPOLYGON (((-86.38406 3...
#> 7 Littleville MULTIPOLYGON (((-87.68859 3...
#> 8 Ragland MULTIPOLYGON (((-86.18473 3...
#> 9 Fort Payne MULTIPOLYGON (((-85.74184 3...
#> 10 Sylvania MULTIPOLYGON (((-85.85684 3...
# 2nd sf in the list:
sf_list$tl_2019_02_place %>% select(NAME, geometry)
#> Simple feature collection with 354 features and 1 field
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -176.6967 ymin: 51.81049 xmax: 173.4299 ymax: 71.34019
#> Geodetic CRS: NAD83
#> First 10 features:
#> NAME geometry
#> 1 Whale Pass MULTIPOLYGON (((-133.1884 5...
#> 2 Utqiagvik MULTIPOLYGON (((-156.9255 7...
#> 3 Anchorage MULTIPOLYGON (((-150.4199 6...
#> 4 Toksook Bay MULTIPOLYGON (((-165.2769 6...
#> 5 Angoon MULTIPOLYGON (((-134.6313 5...
#> 6 Kaktovik MULTIPOLYGON (((-143.6574 7...
#> 7 Point Hope MULTIPOLYGON (((-166.8401 6...
#> 8 Homer MULTIPOLYGON (((-151.655 59...
#> 9 Kachemak MULTIPOLYGON (((-151.4731 5...
#> 10 Kenai MULTIPOLYGON (((-151.3526 6...
Created on 2023-02-05 with reprex v2.0.2
CodePudding user response:
You could define a little function that downloads the zip file, unzips it, reads the shape file into memory, removes the temporary files, then just returns the sf object.
The following function does all that:
read_online_zip_sf <- function(url) {
dir.create("~/zipdir")
f <- tempfile(tmpdir = "~/zipdir", fileext = ".zip")
download.file(url, f)
files <- unzip(f, list = TRUE)
unzip(f, files = files$Name, exdir = "~/zipdir/files")
obj <- sf::st_read("~/zipdir/files")
unlink("~/zipdir", recursive = TRUE)
return(obj)
}
So, now without any mucking about in file explorer, we can do:
url <- paste0("https://github.com/generalpiston/geojson-us-city-boundaries/",
"raw/master/shapes/tl_2019_02_place.zip")
mysf <- read_online_zip_sf(url)
#> Reading layer `tl_2019_02_place' from data source
#> `C:\Users\Administrator\Documents\zipdir\files' using driver `ESRI Shapefile'
#> Simple feature collection with 354 features and 16 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -176.6967 ymin: 51.81049 xmax: 173.4299 ymax: 71.34019
#> Geodetic CRS: NAD83
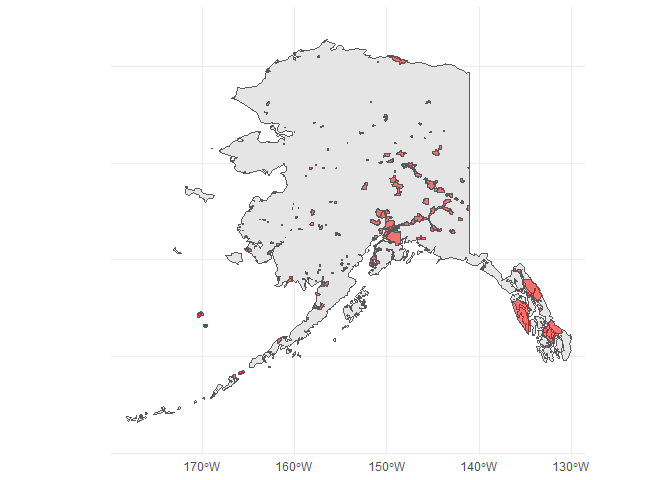
This seems to be a shapefile of city boundaries in Alaska, so let's plot them for completeness:
library(ggplot2)
library(rnaturalearth)
usa <- ne_countries(50, country = "United States of America",
returnclass = "sf")
ggplot(usa)
geom_sf()
geom_sf(data = mysf, fill = "red", alpha = 0.5)
coord_sf(xlim = c(-180, -131), ylim = c(51, 72))
theme_minimal()
Created on 2023-02-05 with reprex v2.0.2
